-
Notifications
You must be signed in to change notification settings - Fork 4
JSON SubstancesProtocols
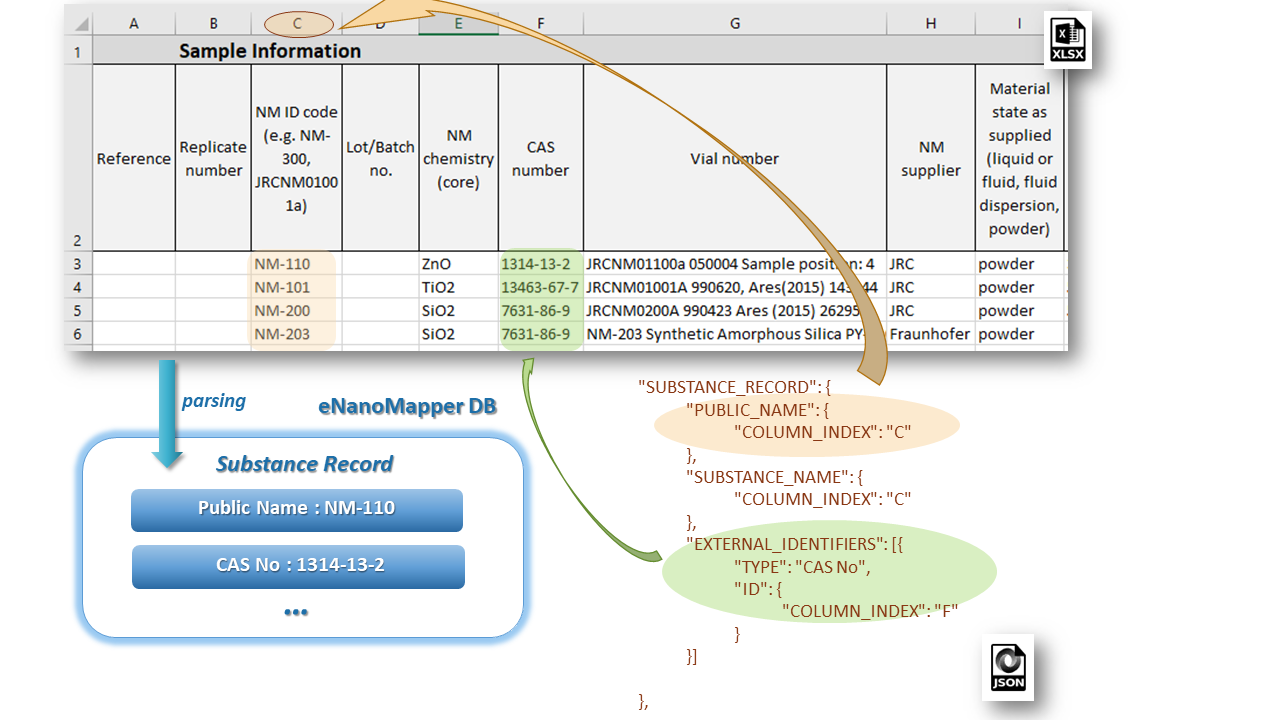
The SUBSTANCE_RECORD section contains a set of EDLs for reading of the basic fields of a Substance Record as described in the data model. This section includes attributes: SUBSTANCE_NAME, OWNER_NAME, SUBSTANCE_TYPE, OWNER_UUID, SUBSTANCE_UUID, PUBLIC_NAME, EXTERNAL_IDENTIFIERS, COMPOSITION, etc. The figure below illustrates the configuration for reading of NM name and CAS number mapped as an external identifier into the eNanoMapper database.
Most often the PUBLIC_NAME field is used for generation of Substance Record UUID which is the basis for many important data operations within eNanoMapper database.
The PROTOCOL_APPLICATIONS section is an array of objects, defining all EDLs concerning Protocol Application data import. Each section includes attributes such as: CITATION_TITLE, CITATION_YEAR, CITATION_OWNER, INTERPRETATION_RESULT, INTERPRETATION_CRITERIA, PROTOCOL_GUIDELINE, PARAMETERS - an array EDLs for the Protocol Application parameters, EFFECTS - an array of sections with EDLs for measurement data and associated metadata, etc.
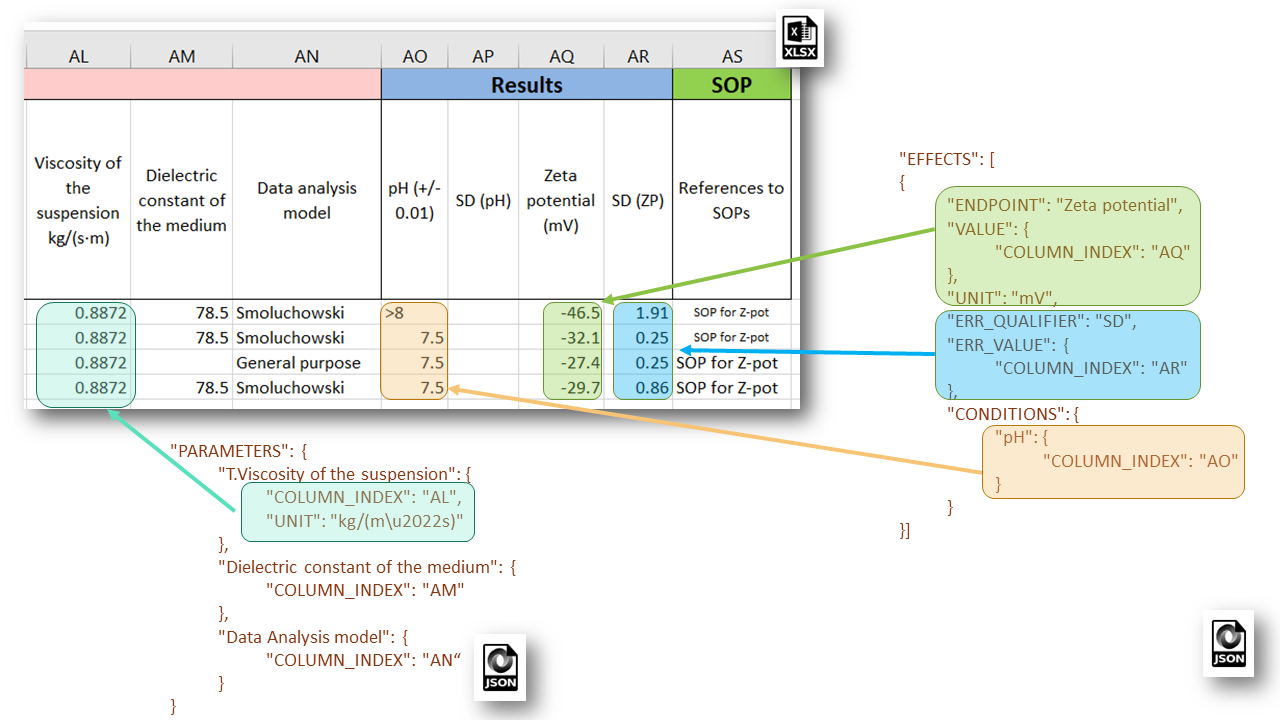
Each EFFECTS section configures the mapping of spreadsheet data onto the Effect Record objects from the eNanoMapper data model, used for storing particular measurements and includes EDL attributes such as: SAMPLE_ID, ENDPOINT, LO_VALUE, UP_VALUE, ERR_VALUE, TEXT_VALUE, VALUE, LO_QUALIFIER, UP_QUALIFIER, ERR_QUALIFIER, UNIT, CONDITIONS (an array of data locations for the experimental conditions). Example JSON configuration of experimental measurement of Zeta potential and corresponding metadata is given in the figure:
eNanoMapper Effect Record is a flexible data structure which allows quite diverse measurement data storage. Each measurement may be a single number or an interval with lo and up values together with specified qualifiers. For example the value of measurement x can be defined like:
x = 3
x < 3
3 < x <= 4
3 < x < 4
x ~ 3
x = 3 +- 0.5
The measurement errors also can be stored with a separate qualifier thus different approaches for setting the measurement uncertainty are supported (s.d. is just one of the possible qualifiers). The measurement value also can be an arbitrary text which allows more complex aggregated data to imported as a string or some nonstandard ways data reporting to be captured. NMDataParser supports an intelligent recognition of the Excel cells content where data, units, intervals, errors and qualifiers may be in a single cell or spread on separate Excel cells. For example: string “< 30 nm” could be in a single cell or “<30” and “nm” in two different cells or “<”, 30 and “nm” in three separate cells. All the latter example cases are fully supported and such flexibility is crucially needed for the purposes of NSC variability in NM experimental data reporting.
- Home
- Quick start
- Data templates
- eNanoMapper Data Model
- Parser configuration via JSON
- Available templates
- How to
- Additional information