-
Notifications
You must be signed in to change notification settings - Fork 4
How conversion works
The conversion relies on a common data model. The input files are parsed into the internal data model. The data can then be exported into several different formats.
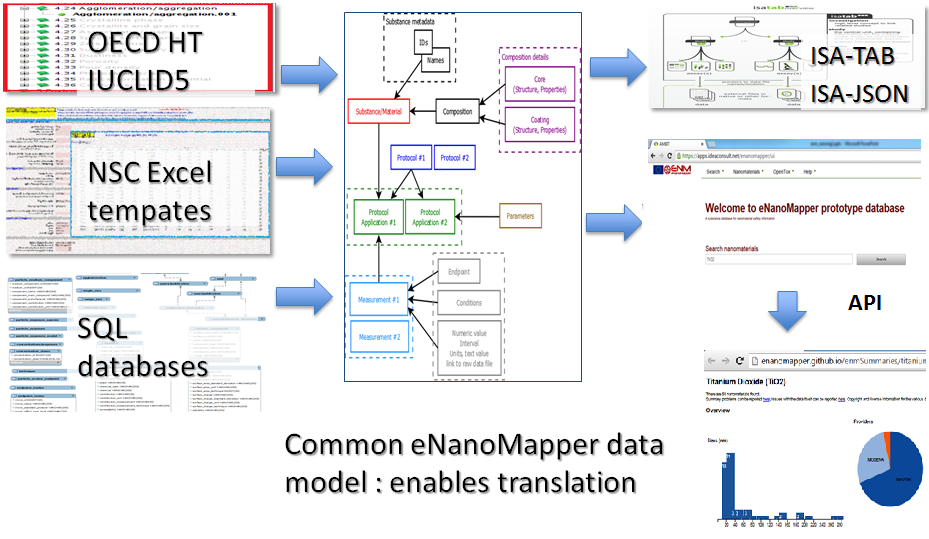
eNanoMapper has developed doi:10.3762/bjnano.6.165 a configurable Excel parser (https://github.com/enanomapper/nmdataparser), enabling import of NanoSafety Cluster Excel templates. The configuration metadata for the parser is defined in a separate file, mapping the custom spreadsheet structure into the internal eNanoMapper storage components. More details on the parser, the configuration file syntax, how to convert between data formats and how to import into the database.
eNanoMapper has developed a working prototype of a utility to export assay data from the eNanoMapper database into ISA-JSON format version 1. Where export to the older ISA-TAB format is needed, ISAtools can be used to convert both to and from the ISA-JSON to ISA-TAB formats. More information on ISA-JSON export.
RDF export following BioAssay Ontology data model is also available. See how to convert between data formats.
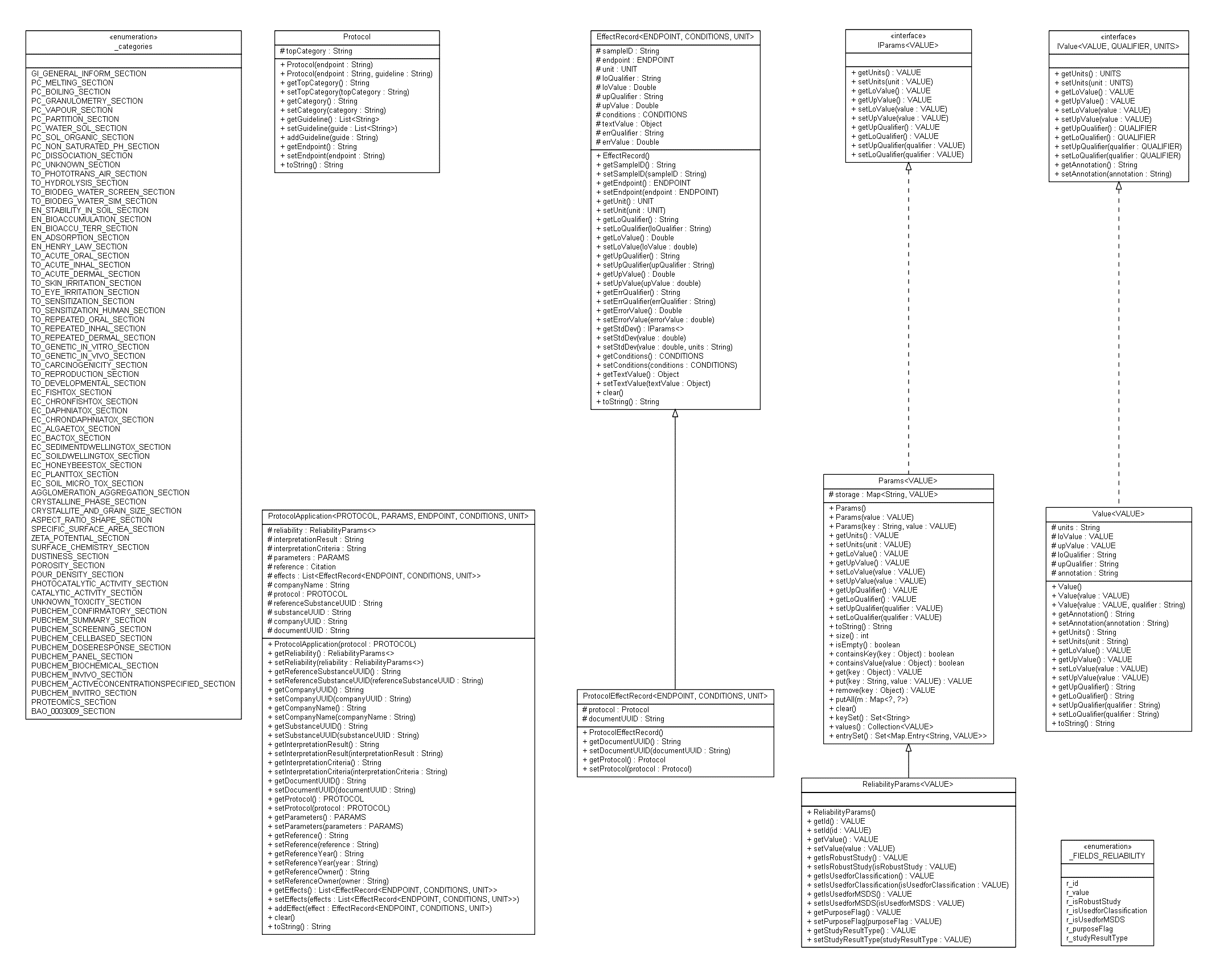
https://enanomapper.adma.ai/datamodel/
Java classes for describing measurements
- Home
- Quick start
- Data templates
- eNanoMapper Data Model
- Parser configuration via JSON
- Available templates
- How to
- Additional information