-
Notifications
You must be signed in to change notification settings - Fork 4
Investigating KAM Network Properties
The Cytoscape Results Panel displays information about KAM Nodes and Edges in your network. This information is available by selecting View -> Show Results Panel from the main menu.
The KAM Node Info tab in the Results Panel displays the supporting BEL Terms for any selected node in the network.
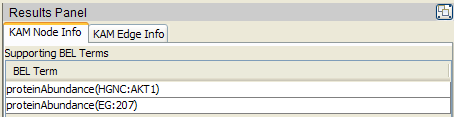
In this example, the p(HGNC:AKT1) node is selected from the Small Corpus (BELScript format) KAM
network. In this KAM, this node is supported by two BEL Terms that were identified as equivalent
during KAM compilation, p(HGNC:AKT1), the protein abundance designated by the value AKT1 in
the HGNC namespace, and p(EG:207), the protein abundance designated by the value 207 in the
EG (Entrez Gene Id) namespace.
The KAM Edge Info tab in the Results Panel displays the supporting BEL Statements for any selected edge in the network, as well as the associated citation and annotation information for each BEL Statement.
images/resultsPanelEdgeNetwork.png
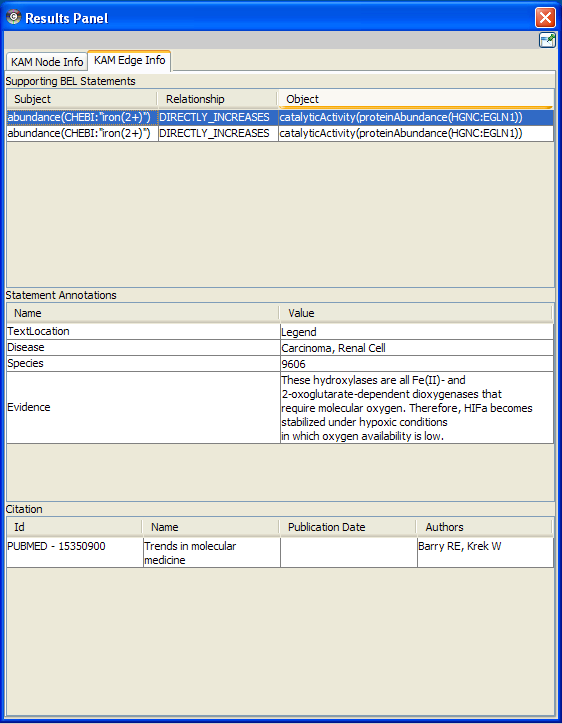
In this example, the selected edge (red) is supported by five statements in the Small Corpus (BELScript format) KAM. The first of the statements is selected, and associated statement annotations and citation displayed.