-
Notifications
You must be signed in to change notification settings - Fork 4
Add nodes by function examples
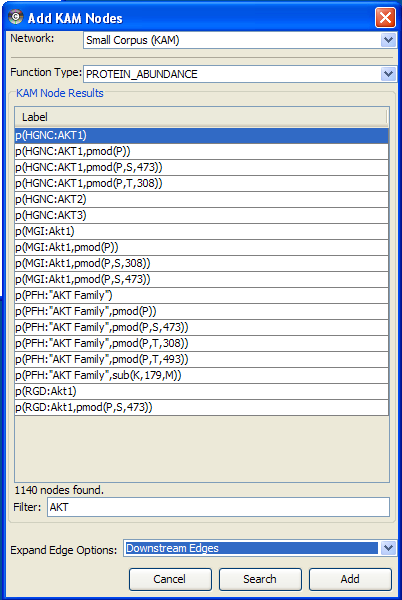
In this example, the Function Type PROTEIN_ABUNDANCE has been selected in the Add KAM Nodes query window, and the results filtered for nodes with labels that contain 'AKT'. The protein abundance of human AKT1 node is selected and the Expand Edge Option Downstream Edges node is enabled. This query will add the human AKT1 node to the network and add any additional nodes and edges that are immediately downstream (i.e., adjacent) to the node. The nodes are added to the KAM Network and displayed in Cytoscape.
The resulting network contains a total of six nodes and five edges, the selected
p(HGNC:AKT1) node, and five downstream nodes. The node selected through
the Add KAM Nodes window is selected (yellow) in the network.
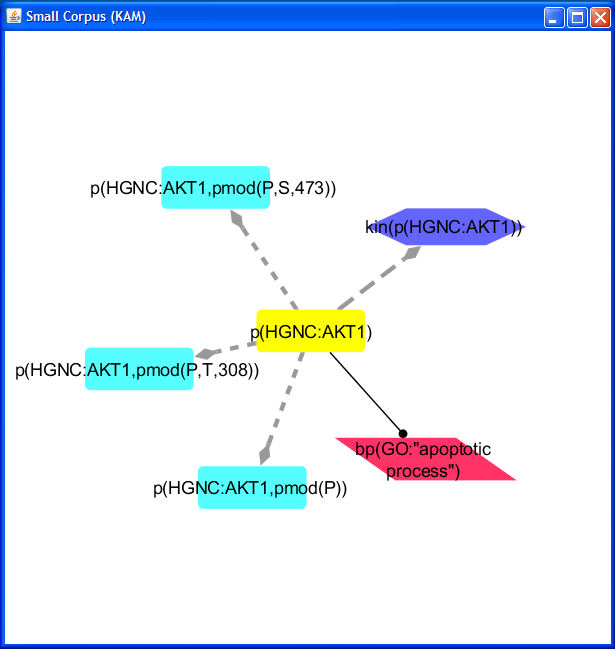
After you add your node(s), you may want to apply a layout; we applied the Cytoscape force-directed layout to all nodes for this example.
In this example, two AKT1 protein abundance nodes are selected from the Add KAM Nodes query window, one with the protein modification phosphorylation at Serine 473, and one with phosphorylation at Threonine 308. All Edges is selected as the Expand Edge Option.
images/addNodesForExample2.png
This query will add two the two modified protein abundance nodes to the network and add any additional nodes and edges upstream or downstream of the two selected nodes.
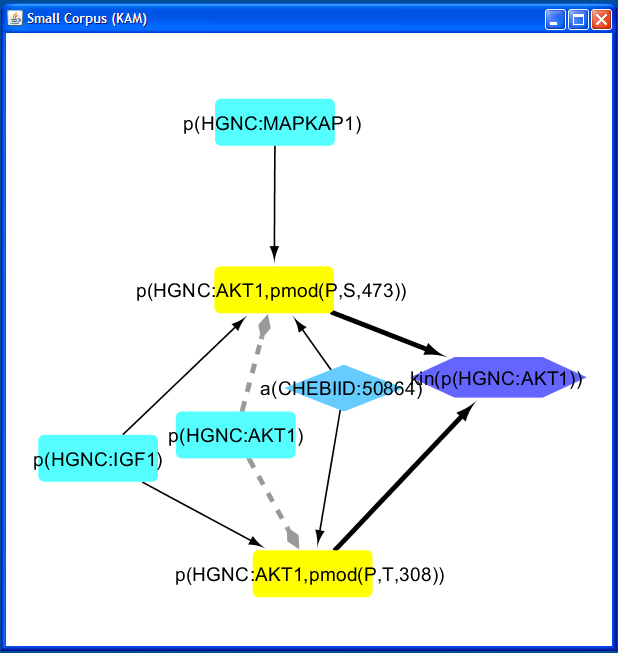
The resulting network contains seven nodes and nine edges, in total. The two selected nodes
(yellow), the upstream nodes a(CHEBIID:50864), p(HGNC:AKT1),
p(HGNC:IGF1), and p(HGNC:MAPKAP1), and the
downstream node kin(p(HGNC:AKT1)).