Fall 2020 semester project (COM-508) carried on at LTS5 at EPFL.
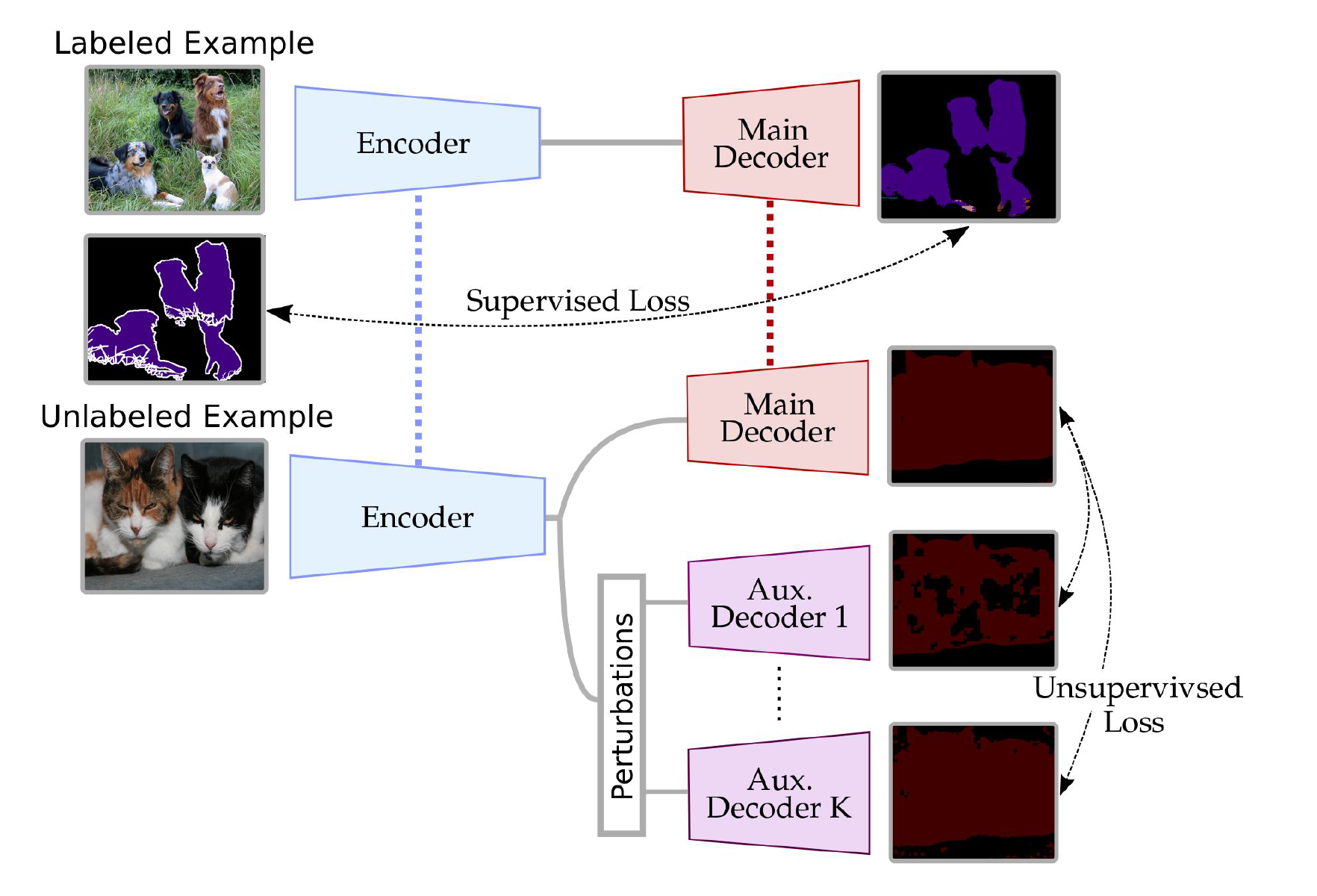
Adapted work of "Semi-supervised semantic segmentation wit cross-consistency training, Ouali et al., 2020". The goal is to reproduce the multiple domain results of their paper using multisite Prostate MRI dataset.
The data used was gathered form multiple challenges by Quande Liu. It contains T2-weighted MRI form six different sites.
Once the data is downloaded it should be organized a followed under the data folder:
- Six folders named : BIDMC, HK , I2CVB, ISBI, ISBI_15 and UCL
- Each subfolders contain two folder : Images and Segmentation
- Each of this folder contains the .nii files (images or mask) for the desired site
To split the data use the file split_data.py which saves the txt files of the split automatically under the right folder.
To train a model :
- Set the config.json file with the desired parameters
- Go to the Prostate_CCT directory and type :
python train.py --config configs/config.json --sites NameName can take different input:
--sites All Call train_multi.py which automatically jointly train the six domain
--sites BIDMC/HK/I2CVB/ISBI/ISBI_15/UCL Call train_single.py which automatically train the desired site alone
The log files and the .pth checkpoints will be saved in saved\EXP_NAME, to monitor the training using tensorboard, please run:
tensorboard --logdir savedTo resume single training using a saved .pth model:
python train.py --config configs/config.json --resume1 saved/CCT/checkpoint.pth --sites BIDMC/HK/I2CVB/ISBI/ISBI_15/UCLTo resume joint training using saved .pth models:
python train.py --config configs/config.json --resume1 saved/CCT/checkpoint.pth --resume2 saved/CCT/checkpoint.pth --resume3 saved/CCT/checkpoint.pth --resume4 saved/CCT/checkpoint.pth --resume5 saved/CCT/checkpoint.pth --resume6 saved/CCT/checkpoint.pth --sites AllThe order of the saved models matter and should be : BIDMC,HK,I2CVB,ISBI,ISBI_15,UCL
Results: The results will be saved in saved as an html file, containing the validation results,
and the name it will take is experim_name specified in configs/config.json. Please do not forget to change the name each time.
For inference, we need a pretrained model, the jpg images we'd like to segment and the config used in training (to load the correct model and other parameters),
python inference.py --config config.json --model best_model.pth --site Name --experiment exp_name --overlay BooleanThe predictions will be saved as .png images in outputs/site/exp_name.
Here are the flags available for inference:
--site Site name of the data to test
--model Path to the trained pth model. (Located in /outputs/EXP_NAME/...)
--config The config file used for training the model. (Located in /outputs/EXP_NAME/...)
--experiment Name of the folder which will contain the results
--overlay If True saves the mri images with an overlay of the segmentation (one for the label and one for the prediction)
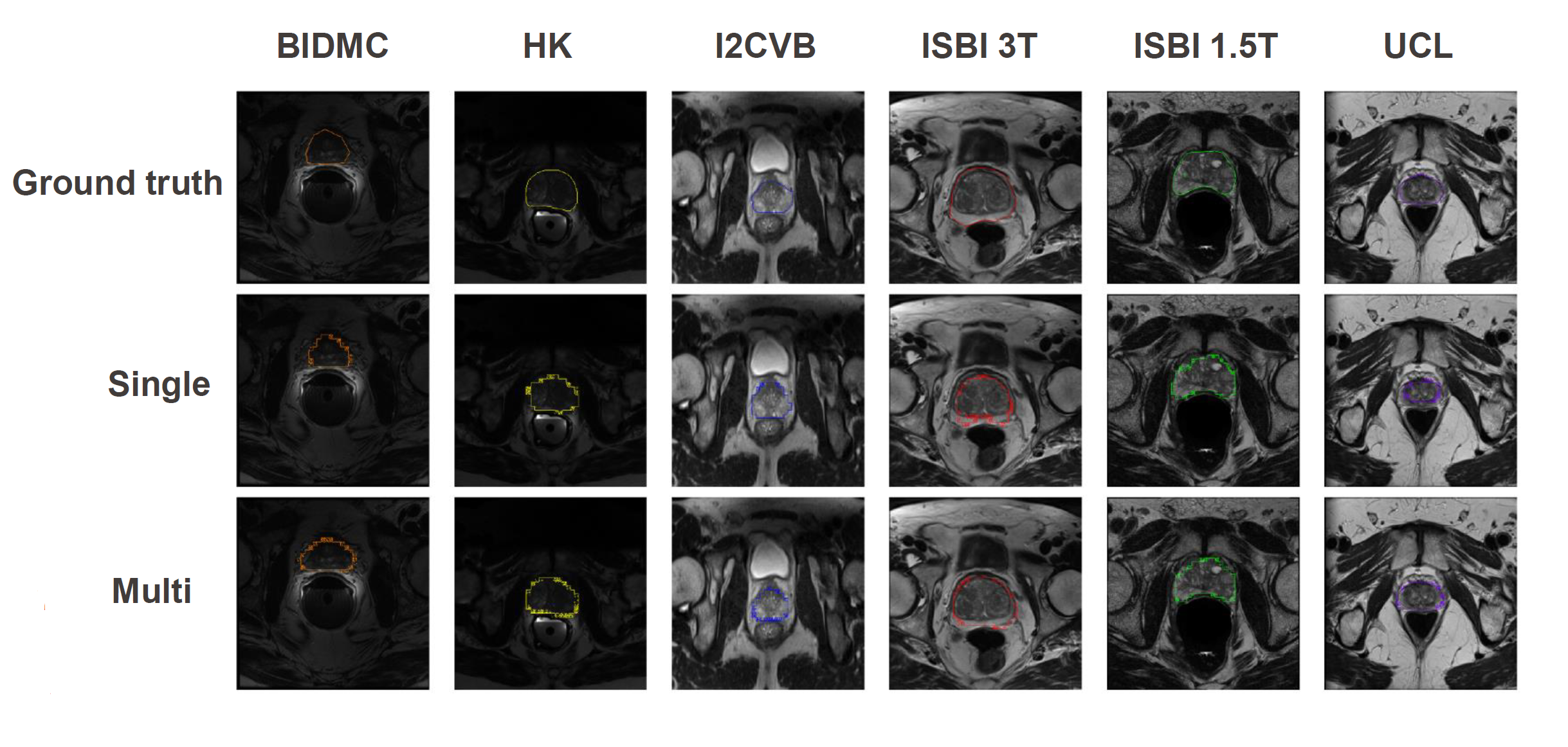
The results are displayed for each domains with the comparison of the ground-truth, single-site training and joint multiple sites training.
Results shown here are not satisfactory, which might be due to the architecture of the segmentation network (PSP-resnet network). We hypothesize that by changing to a segmentation network well-known in medical problem could enhanced the results. The code provide the UNET network, but there has been no time to really explore it.