The official implementation of ACL 2022 paper "Doctor Recommendation in Online Health Forums via Expertise Learning".
Our dataset (avaliable at dataset directory) was collected from Chunyu Yisheng(春雨医生). Our dataset is collected by a crawler within the constraints of the forum. Apart from the personal information de-identified by the forum officially, to prevent privacy leaks, we manually reviewed the collected data and deleted sensitive messages.
embed.csv,train.csv,valid.csv,test.csvtrain.csv,valid.csv,test.csvare the training, validation, testing splits of our dataset respectively.embed.csvis the combination of these three csv files i.e., total data.- they have the same colums:
- "dr_id": doctor ID
- "dialog_id": ID of both dialogues and queries.
- "q": query content
- "parsed_dialog": parsed dialogues (for a dialogue d, we convert it into a token sequence via linking turns in chronological order.)
dialogues.json: dialogues with raw formatdr_profile.jsonl: doctor informations (we use "goodat" of each doctor as profile.)
| # of dialogues | 119,128 |
|---|---|
| # of doctors | 359 |
| # of departments | 14 |
| # of tokens in vocabulary | 8,715 |
| Avg. # of dialogues per doctor | 331.83 |
| Avg. # of doctors per department | 25.64 |
| Avg. # of tokens in a query | 89.97 |
| Avg. # of tokens in a dialogue | 534.28 |
| Avg. # of tokens in a profile | 87.53 |
- Make sure having python libarary
virtualenvinstalled, or install withpip install virtualenv. - Initialize a new virtualenv and install all dependencies.
python -m venv env # create virtual environment source env/bin/activate # activate virtual environment. pip install -r requirements.txt # install all dependencies
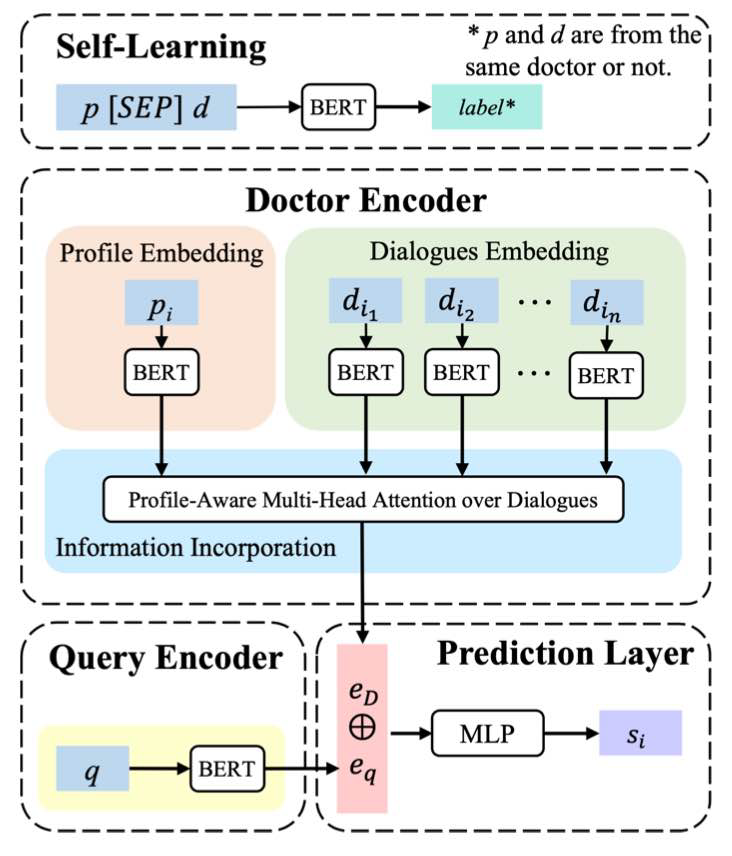
This self-learning task is to predict whether a profile and a dialogue come from the same doctor, where random profile-doctor pairs are adopted as the negative samples. We first fine-tuned mc_bert_base (a pre-trained Chinese Biomedical BERT) via self-learning.
The dataset for self-learning is avaliable at self-learning/dataset. To run both training and evaluation of self-learing task, turn to the self-learning directory, run:
python self_learning.py -seed 2021 -epoch_num 20 -batch_size 50 -accumulation_steps 5Checkpoints will be stored in self_learning/checkpoints directory. We choose our best self-learning checkpoint and move it into 'sl_best_model' that will be used later.
We employ a pre-trained MC-BERT (fine-tuned via self-learning) to encode profile, dialogues, queries and obtain their rudimentary embeddings: dialog_embeddings.json, profile_embeddings.json and q_embeddings.json.
# load self-learning finetuned model from sl_best_model
# output embeddings path: bert_embeddings
python embed.py -load_sl_model 1
# load mc_bert_base model (i.e., without finetuning) from mc_bert_base
# output embeddings path: bert_embeddings_wo_sl
python embed.py -load_sl_model 0 MUL-ATT: With embeddings of doctor profiles, dialogues and queries, it then employs profile-aware multi-head attention over dialogues to explore doctor expertise and works with the query encoder (to capture patient needs) to pair doctors with queries.
Recommendation Prediction: Given a pair of doctor
We provide three bashs scripts train.sh, test.sh and eval.sh to run the training, prediction, and evaluation of three MUL-ATT models:
- MUL-ATT (W/O SL): multi-head attention without this self-learning step
- MUL-ATT (W/O D): encode profiles only with a multi-head self-attention
- MUL-ATT (W/O P): with dialogues only
- MUL-ATT (FULL): the full model
train.sh and predict.sh will call python train.py and python predict.py respectively for training and prediction. The experiment settings in train.sh and predict.sh are corresponding to settings stated in config.py.
Note that all experiments will be run in parallel and select a single available GPU sequentially. You can change the number of total gpus (i.e. n_gpu) in
train.shandpredict.sh. To monitor experiments, you may view the corresponding generated log file.
For the evaluation (i.e., eval.sh), we use RankLib to evaluate the predictions with information retrieval metrics: precision@N (P@N), mean average precision (MAP), and ERR@N . N is set to 1 for P@N and 5 for ERR@N.
@inproceedings{lu-etal-2022-doctor,
title = "Doctor Recommendation in Online Health Forums via Expertise Learning",
author = "Lu, Xiaoxin and
Zhang, Yubo and
Li, Jing and
Zong, Shi",
booktitle = "Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers)",
month = may,
year = "2022",
address = "Dublin, Ireland",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/2022.acl-long.79",
pages = "1111--1123",
}