-
Notifications
You must be signed in to change notification settings - Fork 4
Module: TransmissionTimeSample
Niema Moshiri edited this page Jul 9, 2018
·
65 revisions
The TransmissionTimeSample module chooses the time at which a given transmission event will occur. See the source code to see what is defined by the abstract class.
- Each transmission event occurs after some time delta after the previous transmission event, where delta is sampled from an exponential distribution with a user-specified rate
- Requirements:
- Config Parameters:
-
time_sample_rate: Rate of the exponential distribution from which to sample time deltas
-
- Each transmission event occurs a fixed time delta after the previous transmission event
- Requirements:
- None
- Config Parameters:
-
fixed_transmission_time_delta: Fixed time delta between consecutive transmission events
-
- Transmission network is simulated the transmission model developed by Hethcote and Yorke as it is described in this paper using GEMF
- The states are Male and Female Abstinent (MA and FA), Male and Female Susceptible (MS and FS), Male and Female Symptomatic Infected (MIS and FIS), and Male and Female Asymptomatic Infected (MIA and FIA)
-
 * We assume the MA and FA states are uninfected (i.e., individuals who are infected and transition to abstinence are cured as well)
* We assume the MA and FA states are uninfected (i.e., individuals who are infected and transition to abstinence are cured as well)
- Requirements:
- Must use EndCriteria_GEMF module
- Must use TransmissionNodeSample_GEMF module
- Nodes in the contact network must have an attribute of either
MALEorFEMALE- See the Contact Network File Format documentation
- Config Parameters:
-
gemf_path: The path to yourGEMFexecutable (or simply"GEMF"if it is in yourPATHvariable) -
end_time: The time at which to end the transmission simulation - All transition rates (see entry in FAVITES_ModuleList.json for complete list)
-
- Transmission network is simulated the HIV-ART transmission model as it is described in this paper using GEMF
- The states are Non-Susceptible (NS), Susceptible (S), four stages of Infected (I1, I2, I3, and I4), four stages of ART (A1, A2, A3, and A4), and Deceased (D)
-

- Requirements:
- Must use EndCriteria_GEMF module
- Must use TransmissionNodeSample_GEMF module
- Config Parameters:
-
gemf_path: The path to yourGEMFexecutable (or simply"GEMF"if it is in yourPATHvariable) -
end_time: The time at which to end the transmission simulation - All transition rates (see entry in FAVITES_ModuleList.json for complete list)
-
- Transmission network is simulated the HIV HPTN 071 (PopART) transmission model as it is described in this paper using GEMF
- The following states exist in the model:
- Male Susceptible Uncircumcised (MSU), Pending Circumcision (MSPC), Circumcised and Healing (MSCH), Circumcised (MSC), and Female Susceptible (FS)
- Male and Female Acute Infection (MIA and FIA), Infected with CD4≥500 (MI1 and FI1), 350≤CD4<500 (MI2 and FI2), 200≤CD4<350 (MI3 and FI3), and CD4<200 (MI4 and FI4)
- Male and Female Infected with Treatment Pending with CD4≥500 (MJ1 and FJ1), 350≤CD4<500 (MJ2 and FJ2), 200≤CD4<350 (MJ3 and FJ3), and CD4<200 (MJ4 and FJ4)
- Male and Female Infected with Treatment but Not Virally Suppressed with CD4≥500 (MT1 and FT1), 350≤CD4<500 (MT2 and FT2), 200≤CD4<350 (MT3 and FT3), and CD4<200 (MT4 and FT4)
- Male and Female Infected with Treatment and Virally Suppressed with CD4≥500 (MA1 and FA1), 350≤CD4<500 (MA2 and FA2), 200≤CD4<350 (MA3 and FA3), and CD4<200 (MA4 and FA4)
- Deceased (D), to which all other states have an edge
-

- The following states exist in the model:
- Requirements:
- Must use EndCriteria_GEMF module
- Must use TransmissionNodeSample_GEMF module
- Config Parameters:
-
gemf_path: The path to yourGEMFexecutable (or simply"GEMF"if it is in yourPATHvariable) -
end_time: The time at which to end the transmission simulation - All transition rates (see entry in FAVITES_ModuleList.json for complete list)
-
- Each transmission occurs at the current global time
- Requirements:
- None
- Config Parameters:
- None
- Wrapper for PANGEA.HIV.sim
- This is not supported in the Docker/Singularity images from FAVITES 1.1.11 onward
- Requirements:
- R
- PANGEA.HIV.sim
- Must use ContactNetwork_PANGEA module
- Must use ContactNetworkGenerator_PANGEA module
- Must use EndCriteria_Instant module
- Must use NodeEvolution_PANGEA module
- Must use NodeSample_PANGEA module
- Must use NumBranchSample_All module
- Must use NumTimeSample_PANGEA module
- Must use PostValidation_Dummy module
- Must use SeedSelection_PANGEA module
- Must use SeedSequence_PANGEA module
- Must use SequenceEvolution_PANGEA module
- Must use SourceSample_PANGEA module
- Must use TimeSample_PANGEA module
- Must use TransmissionNodeSample_PANGEA module
- Must use TransmissionTimeSample_PANGEA module
- Must use TreeUnit_Same module
- Config Parameters
-
Rscript_path: The path to yourRscriptexecutable (or simply"Rscript"if it is in yourPATHvariable) - All
pangea_parameters, which correspond to PANGEA.HIV.sim parameters (see entry in FAVITES_ModuleList.json for complete list, and see PANGEA.HIV.sim help for details)- Use
""for default
- Use
-
- Transmission network is simulated via the SAIS model using GEMF
-
- Requirements:
- Must use EndCriteria_GEMF module
- Must use TransmissionNodeSample_GEMF module
- Config Parameters:
-
gemf_path: The path to yourGEMFexecutable (or simply"GEMF"if it is in yourPATHvariable) -
end_time: The time at which to end the transmission simulation -
sais_beta_seed: The desired susceptible infection rate (S→I) from outside the contact network (i.e., seed infection) -
sais_beta_by_i: The desired susceptible infection rate (S→I) from within the contact network -
sais_beta_a_seed: The desired aware infection rate (A→I) from outside the contact network (i.e., seed infection) -
sais_beta_a_by_i: The desired aware infection rate (A→I) from within the contact network -
sais_kappa: The desired alert rate (S→A) -
sais_delta: The desired recovery rate (I→S)
-
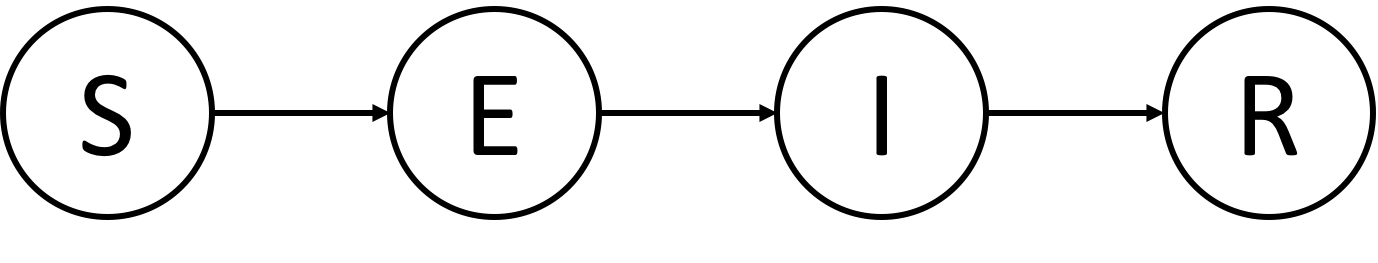
- Transmission network is simulated via the SEIR model using GEMF
-
- Requirements:
- Must use EndCriteria_GEMF module
- Must use TransmissionNodeSample_GEMF module
- Config Parameters:
-
gemf_path: The path to yourGEMFexecutable (or simply"GEMF"if it is in yourPATHvariable) -
end_time: The time at which to end the transmission simulation -
seir_beta_seed: The desired infection rate (S→E) from outside the contact network (i.e., seed infection) -
seir_beta_by_i: The desired infection rate (S→E) from within the contact network -
seir_lambda: The desired rate of becoming infectious (E→I) -
seir_delta: The desired recovery rate (I→R)
-
- Transmission network is simulated via the SI model
-
- Requirements:
- Must use
TransmissionNodeSample_SImodule
- Must use
- Config Parameters:
-
infection_rate: The rate at which susceptible individuals are infected (per infected and time)
-
- Transmission network is simulated via the SI model using GEMF
-
- Requirements:
- Must use EndCriteria_GEMF module
- Must use TransmissionNodeSample_GEMF module
- Config Parameters:
-
gemf_path: The path to yourGEMFexecutable (or simply"GEMF"if it is in yourPATHvariable) -
si_beta_seed: The desired infection rate (S→I) from outside the contact network (i.e., seed infection) -
si_beta_by_i: The desired infection rate (S→I) from within the contact network -
end_time: The time at which to end the transmission simulation
-
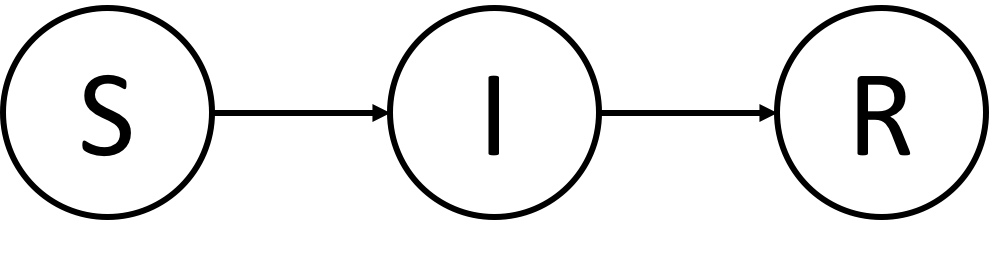
- Transmission network is simulated via the SIR model using GEMF
-
- Requirements:
- Must use EndCriteria_GEMF module
- Must use TransmissionNodeSample_GEMF module
- Config Parameters:
-
gemf_path: The path to yourGEMFexecutable (or simply"GEMF"if it is in yourPATHvariable) -
end_time: The time at which to end the transmission simulation -
sir_beta_seed: The desired infection rate (S→I) from outside the contact network (i.e., seed infection) -
sir_beta_by_i: The desired infection rate (S→I) from within the contact network -
sir_delta: The desired recovery rate (I→R)
-
- Transmission network is simulated via the SIS model using GEMF
-
- Requirements:
- Must use EndCriteria_GEMF module
- Must use TransmissionNodeSample_GEMF module
- Config Parameters:
-
gemf_path: The path to yourGEMFexecutable (or simply"GEMF"if it is in yourPATHvariable) -
end_time: The time at which to end the transmission simulation -
sis_beta_seed: The desired infection rate (S→I) from outside the contact network (i.e., seed infection) -
sis_beta_by_i: The desired infection rate (S→I) from within the contact network -
sis_delta: The desired curing rate (I→S)
-
- Transmission network is simulated via an extension of the SIR model dubbed "SVITR" (Susceptible, Vaccinated, Infected, Treated, Recovered) using GEMF
-
- Requirements:
- Must use EndCriteria_GEMF module
- Must use TransmissionNodeSample_GEMF module
- Config Parameters:
-
gemf_path: The path to yourGEMFexecutable (or simply"GEMF"if it is in yourPATHvariable) -
end_time: The time at which to end the transmission simulation -
svitr_s_to_v: The desired vaccination rate (S→V) -
svitr_beta_seed: The desired infection rate (S→I) caused by nodes outside the contact network -
svitr_beta_by_i: The desired infection rate (S→I) caused by nodes in state I -
svitr_beta_by_t: The desired infection rate (S→I) caused by nodes in state T -
svitr_i_to_t: The desired treatment rate (I→T) -
svitr_delta: The desired untreated recovery rate (I→R) -
svitr_t_to_r: The desired treated recovery rate (T→R)
-
- Transmission network is read from file
- Requirements:
- Must use
EndCriteria_TransmissionFilemodule - Must use
SeedSelection_TransmissionFilemodule - Must use
TransmissionNodeSample_TransmissionFilemodule
- Must use
- Config Parameters:
-
transmission_network_file: The desired transmission network, represented in the FAVITES Transmission Network File Format
-
Niema Moshiri & Siavash Mirarab 2016