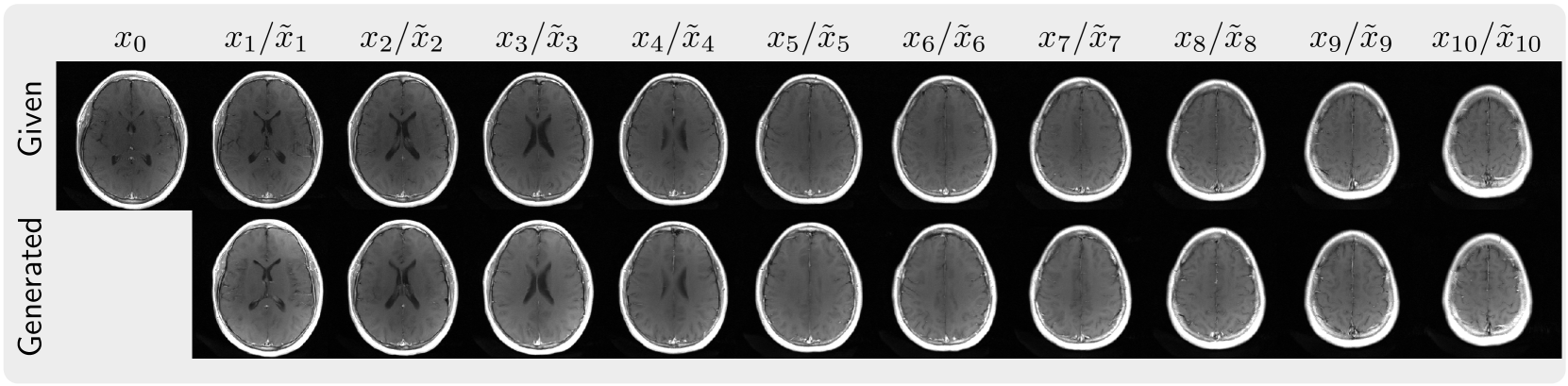
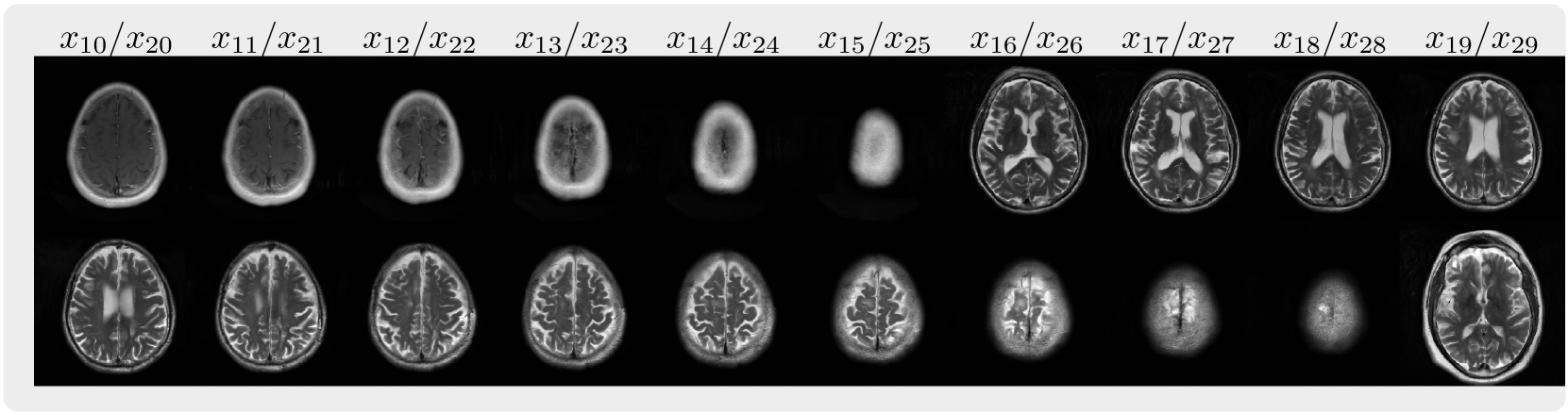
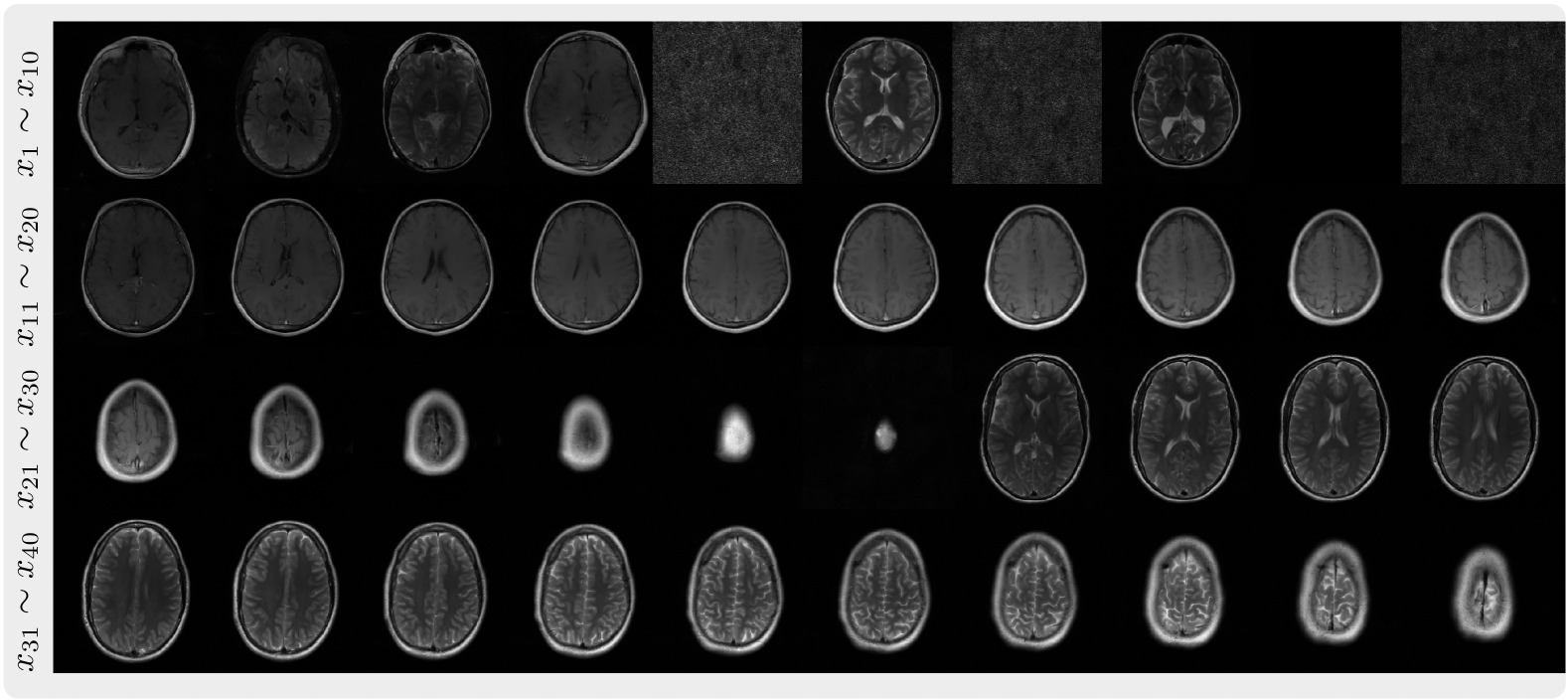
This is the codebase for Autoregressive Image Diffusion (AID), which is built upon openai/guided-diffusion with modifications for the training and sampling of autoregressive image diffusion models and its application to MRI reconstruction.
The Causal-Unet is implemented upon the original Unet architecture to allow for autoregressive mechanisms imposed on image sequences. In gaussian_diffusion.py, we added function training_losses_temporal for training AID models to exploit dependencies in image sequences and some other relevant modifications, and we added function p_sample_temporal_loop for sampling image sequences prospectively and some other relevant modifications.
| FastMRI 2D multi-slices | Cardiac Cine | ABIDE 3D volume |

|

|

|
git clone https://github.com/mrirecon/aid.git
cd aid
pip install -e .cd scripts
bash examples.sh
python viewer.py logs/example_xxx/sampleThe results will be saved in scripts/logs/example_xxx, where xxx denotes the experiment name. Please use the viewer.py to visualize the samples. It take around 3 hours to complete the following five tasks in the examples.sh.
download_pretrained: Downloads pretrained models and sample data necessary for reconstruction and sampling.unfolding: Performs MRI unfolding using pretrained models to generate high-resolution images usingsample.py.reconstruction: Conducts volume reconstruction of MRI images using pretrained models usingfastmri_recon.py.sampling_brain: Generates sample MRI images using pretrained models usingsample.py.sampling_cardiac: Generates sample cardiac cine images using pretrained models usingsample.py.
The CustomDataLoader and CustomDataset classes are implemented for loading image sequences for training without breaking the order of the sequences. The function load_data returns a DataLoader object that can be used for training. Please refer to the datasets/README.md for instructions on preparing datasets for training. Update the script train.sh variables according to your environment:
logdir: Directory to save logs and checkpoints.expname: Experiment name for logging and checkpoint saving.datadir: Directory containing the dataset.image_size: Size of the images.
Perform two-stage training manually using the two functions: first_stage and second_stage, in the script train.sh. The second_stage function resumes training from the first stage checkpoint and only trains the temporal-spatial conditioning block. The --resume_checkpoint flag can be used to resume training from a checkpoint.
The one_stage function is a combined training stage with a different configuration suitable for single-stage training.
Pretrained models are hosted on huggingface.
We provided the binary file for BART reconstruction toolbox on this link. If it doesn't work on your local system, please clone the BART repository and compile it on your local system.
We provided a latent space model for cardiac cine image generation, in which the VQVAE model is trained on the cardiac cine dataset using the code in this repository taming-transformers. Please install it when you want to generate cardiac cine images.
We provided recon_all.sh for performing MRI reconstruction of the validation data in the fastMRI dataset on HPC cluster. This script will submit a job for each of the validation data. The job contains many tasks that are defined in recon_func.sh. The script recon_func.sh will perform reconstruction experiments using the pretrained models and save the results in the specified directory. If you are interested in running the script, please update the variables in the script according to your environment.
We provided image_train.py for training the normal image diffusion model. The command to use it is included in train.sh.
If you have any questions, please contact Guanxiong Luo ([email protected]) or raise an issue in the repository.
If you find this code useful, please consider citing the following paper:
@article{luo2024autoregressive,
title={Autoregressive Image Diffusion: Generation of Image Sequence and Application in MRI},
author={Luo, Guanxiong and Huang, Shoujin and Uecker, Martin},
journal={arXiv preprint arXiv:2405.14327},
year={2024}
}