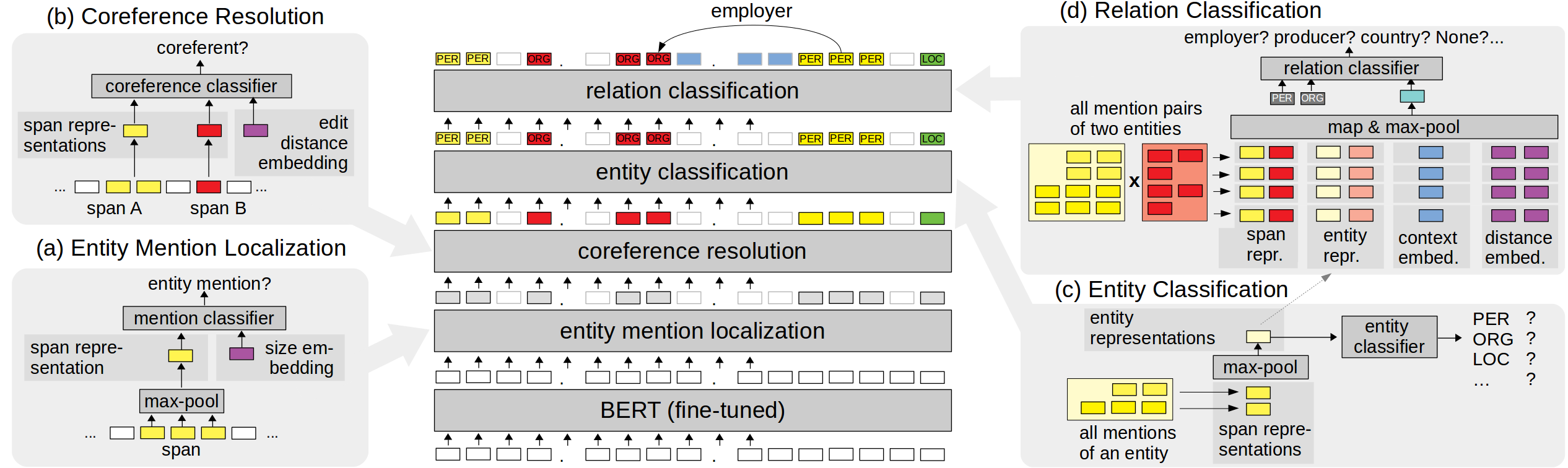
PyTorch code for JEREX: "Joint Entity-Level Relation Extractor". For a description of the model and experiments, see our paper "An End-to-end Model for Entity-level Relation Extraction using Multi-instance Learning": https://arxiv.org/abs/2102.05980 (accepted at EACL 2021).
- Required
- Python 3.7+
- PyTorch (tested with version 1.8.1 - see here on how to install the correct version)
- PyTorch Lightning (tested with version 1.2.7)
- transformers (+sentencepiece, e.g. with 'pip install transformers[sentencepiece]', tested with version 4.5.1)
- hydra-core (tested with version 1.0.6)
- scikit-learn (tested with version 0.21.3)
- tqdm (tested with version 4.43.0)
- numpy (tested with version 1.18.1)
- jinja2 (tested with version 2.11.3)
Execute the following steps before running the examples.
(1) Fetch end-to-end (joint) DocRED [1] dataset split. For the original DocRED split, see https://github.com/thunlp/DocRED :
bash ./scripts/fetch_datasets.sh
(2) Fetch model checkpoints (joint multi-instance model (end-to-end split) and relation classification multi-instance model (original split)):
bash ./scripts/fetch_models.sh
(1) Train JEREX (joint model) using the end-to-end split:
python ./jerex_train.py --config-path configs/docred_joint
(2) Evaluate JEREX (joint model) on the end-to-end split (you need to fetch the model first):
python ./jerex_test.py --config-path configs/docred_joint
To run these examples, first download the original DocRED dataset into './data/datasets/docred/' (see 'https://github.com/thunlp/DocRED' for instructions)
(1) Train JEREX (multi-instance relation classification component) using the orignal DocRED dataset.
python ./jerex_train.py --config-path configs/docred
(2) Evaluate JEREX (multi-instance relation classification component) on the original DocRED test set (you need to fetch the model first):
python ./jerex_test.py --config-path configs/docred
Since the original test set labels are hidden, the code will output an F1 score of 0. A 'predictions.json' file is saved, which can be used to retrieve test set metrics by uploading it to the DocRED CodaLab challenge (see https://github.com/thunlp/DocRED)
- If you want to compare your end-to-end model to JEREX using the strict evaluation setting, have a look at our evaluation script.
- The DocRED dataset contains some duplicate annotations (especially entity mentions). Duplicates are removed during evaluation (i.e. only counted once).
- The hyperparameters used in our paper are set as default. You can adjust hyperparameters and other configuration settings in the 'train.yaml' and 'test.yaml' under ./configs
- Settings can also be overriden via command line, e.g.:
python ./jerex_train.py training.max_epochs=40
- A brief explanation of available configuration settings can be found in './configs.py'
- Besides the main JEREX model ('joint_multi_instance') and the 'global' baseline ('joint_global') you can also train each sub-component ('mention_localization', 'coreference_resolution', 'entity_classification', 'relation_classification_multi_instance', 'relation_classification_global') individually. Just set 'model.model_type' accordingly (e.g. 'model.model_type: joint_global')
- When testing a model ('./jerex_test.py') or by either specifying a test dataset (using 'datasets.test_path' configuration) or setting 'final_valid_evaluate' to True (using 'misc.final_valid_evaluate=true' configuration) during training ('./jerex_train.py'), a file containing the model's predictions is stored ('predictions.json').
- By using a joint model ('joint_multi_instance' / 'joint_global'), a file ('examples.html') containing visualizations of all prediction results is also stored alongside 'predictions.json'.
Performing a search over token spans (and pairs of spans) in the input document (as in JEREX) can be quite (CPU/GPU) memory demanding. If you run into memory issues (i.e. crashing of training/inference), these settings may help:
- 'training.max_spans'/'training.max_coref_pairs'/'training.max_rel_pairs' (or 'inference.max_spans'/'inference.max_coref_pairs'/'inference.max_rel_pairs'): These settings restrict the number of spans/mention pairs for coreference resolution/mention pairs for MI relation classification that are processed simultaneously. Setting these to a lower number reduces training/inference speed, but lowers memory consumption.
- The default setting of maximum span size is quite large. If the entity mentions in your dataset are usually shorter than 10 tokens, you can restrict the span search to less tokens (by setting 'sampling.max_span_size')
[1] Yuan Yao, Deming Ye, Peng Li, Xu Han, Yankai Lin,Zhenghao Liu, Zhiyuan Liu, Lixin Huang, Jie Zhou,and Maosong Sun. 2019. DocRED: A Large-Scale Document-Level Relation Extraction Dataset. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, pages 764–777, Florence, Italy. ACL.