This is a re-implementation of our study "Automated analysis and detection of abnormalities in transaxial anatomical cardiovascular magnetic resonance images: a proof of concept study with potential to optimize image acquisition" in the The International Journal of Cardiovascular Imaging.
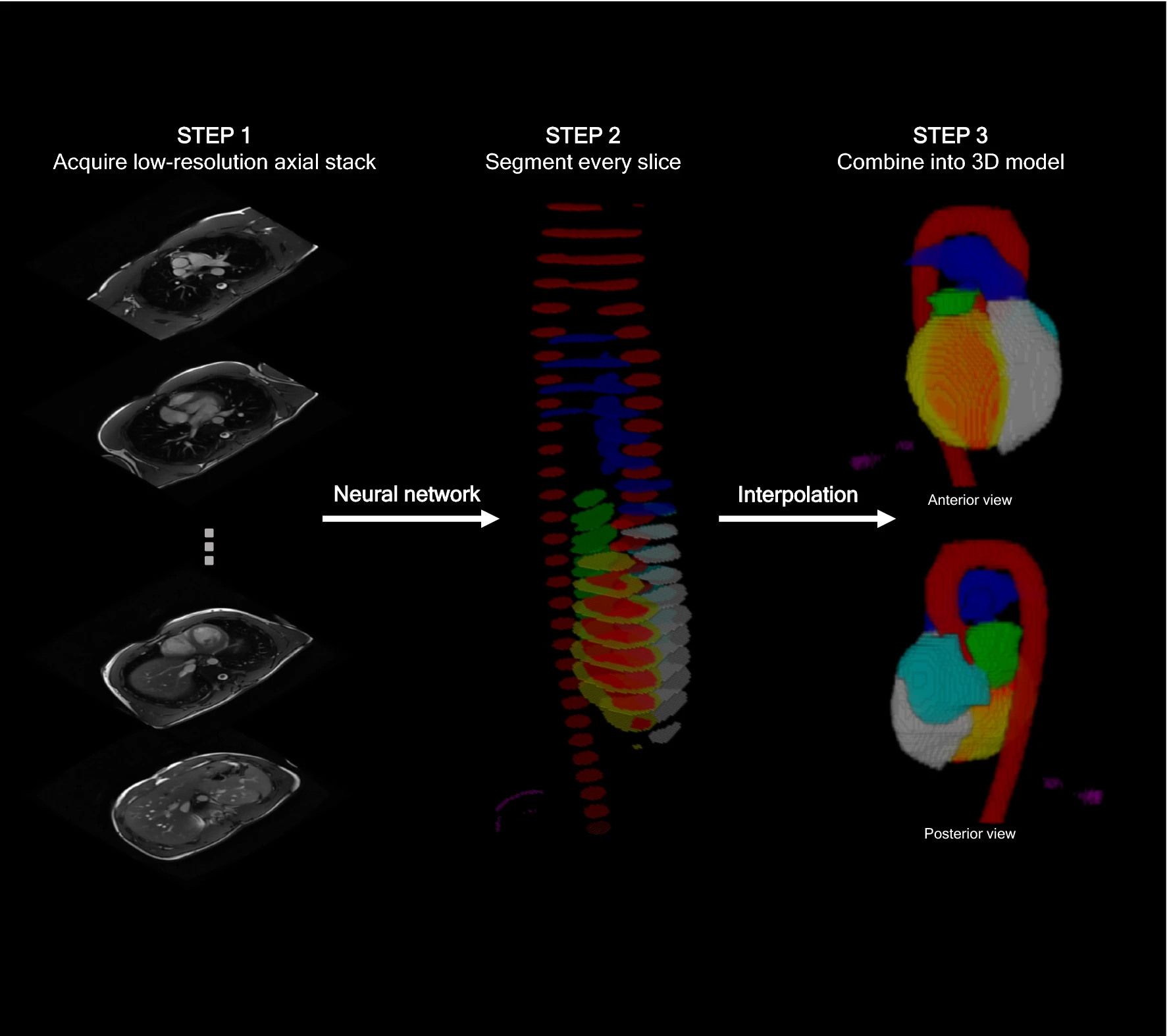
Within the first couple of minutes of a CMR scan, a series of transaxial images are routinely acquired, commonly termed the “anatomy” sequences.
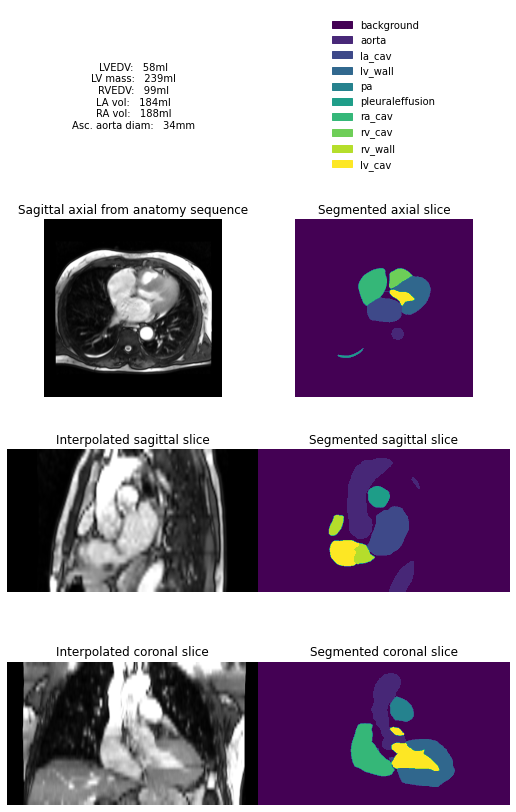
Here we use a neural network to analyse and segment each slice of these images, before combining these slices into a 3D model. This 3D model is then used to estimate common cardiac measurements made by expert humans. The hope is we can get good estimates of these measures right at the start of the scan, so any unexpected findings can be flagged up early.
The Onnx trained model used for inference is available here for CPU and here for CUDA
inference.pynb shows an example where a folder containing a typical anatomy sequence is supplied to the model:
The following files are also available in the repository:
-
1_train.py- Training code to train the Pytorch model -
2_export_onnx.py- Code to export the Onnx model (link to above) -
3_inference_on_all_studies.py- Batch inference on a series of studies, with results exported as a CSV file. -
4_compare_predictions_to_groundtruth.py- The results from Step 3 can be plotted as a scatter plot versus gold standard predictions.