Version: 0.1 (Please note that this is proof-of-concept code in a very early development stage). For qestions and remarks pleaes contact Gerrit Großmann.
Official code for our paper Learning Vaccine Allocation from Simulations.
Install rust:
sudo curl https://sh.rustup.rs -sSf | sh -s -- -y && source $HOME/.cargo/envCompile rust code:
cd rust_code/rust_reject && cargo build --releaseInstall Python3 dependencies (or use pipenv):
pip install -r requirements.txtRun the experiments from the study with:
python evaluation.pyAlternativly, you can import and call def analysis from evaluation.py.
You can also call the rust code diretly.
See the def call_rust for more information.
Note that the network file contains containing a labeled graph specifying the initial state, each line having the form <Nodeid>;<Label>;<Neighbor1>,<Neighbor2>,...
0;I;31,29,94,13,83
1;S;66,15,73
2;S;29,61,26,80,16,83,30,62,3,93,27,87,68,18,79,6
3;I;83,2,29,4,28,61,46,21,9,49,41,68,16,74
4;S;82,28,12,83,3,62,66,68
...Nodes start with id 0 and are sorted (rendering the first number somewhat redundant). Isolates (nodes withouth neighbors) are not supported (yet). There should be at least one node for each possible label.
Each rust-call produces extensive outputs in output/<expname> containing the input graphs, the transmission graphs, and its solution.
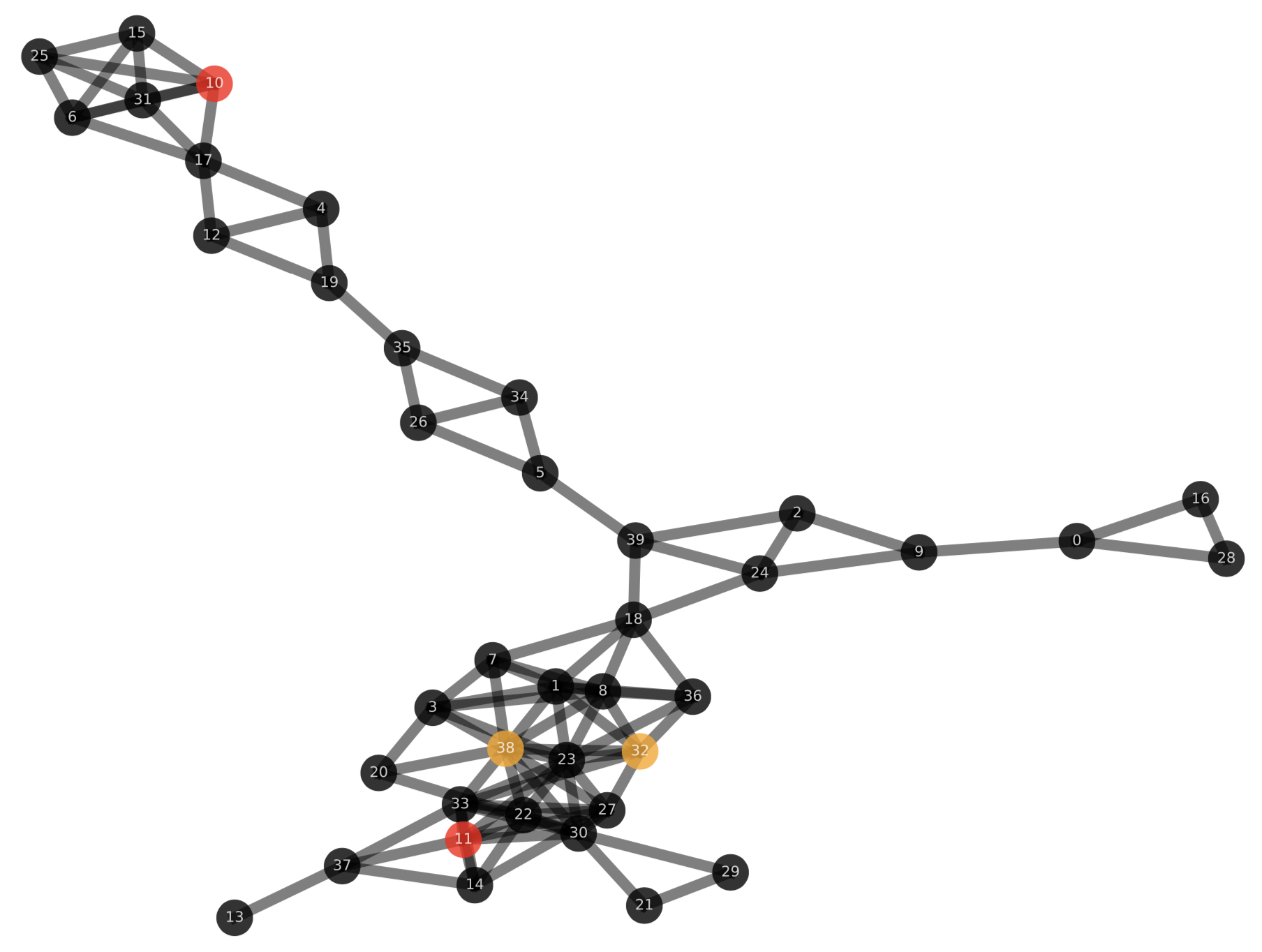
In particular, in each iteration, Simba outputs the corresponding input graph where initially infected nodes are red and vaccinated nodes are orange.
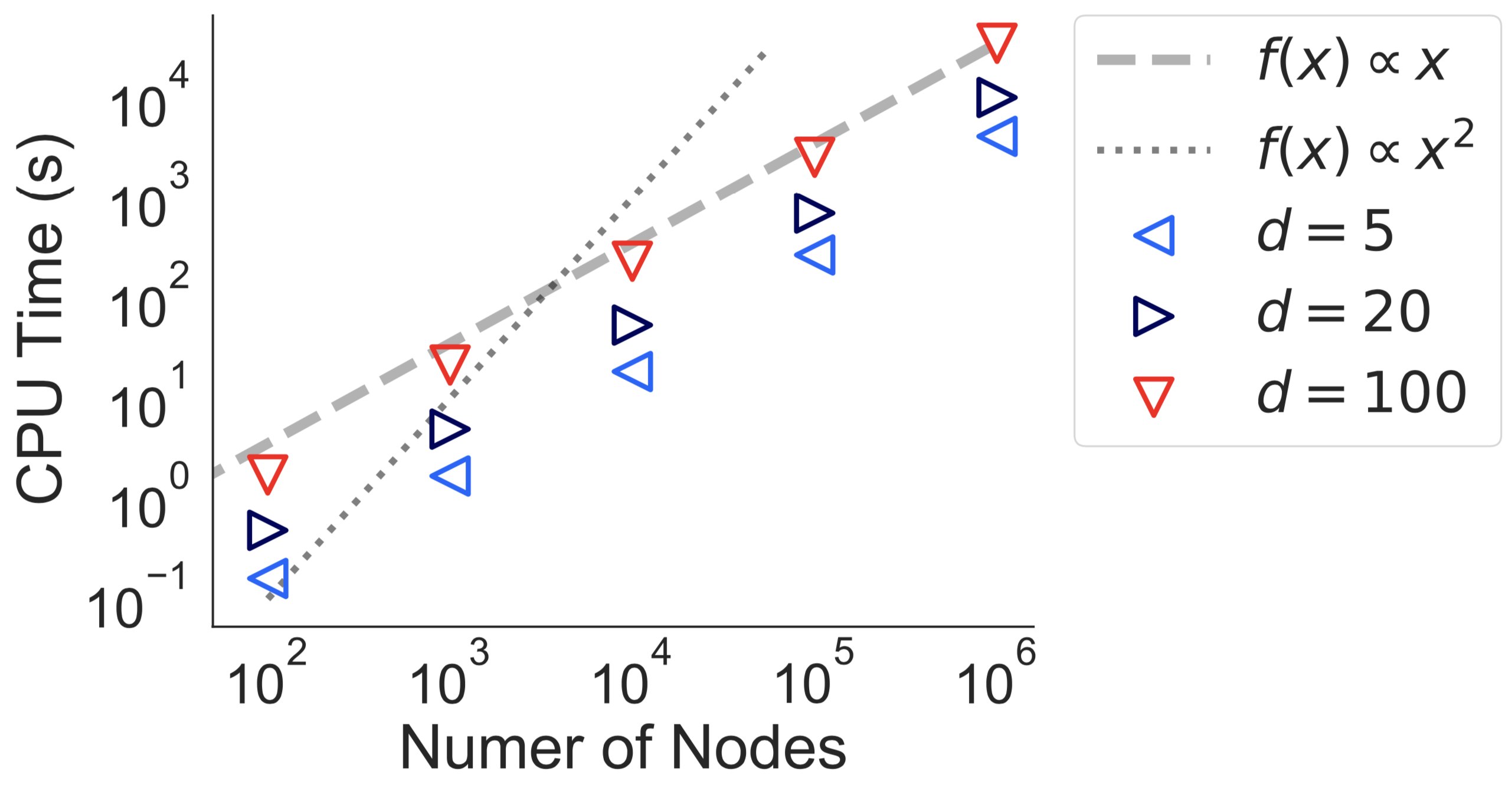
Constructing and solving a single transmission graph is slightly supra-linear.
- Clean output (and direct to log) and code
- Use variance reduction technique in rust