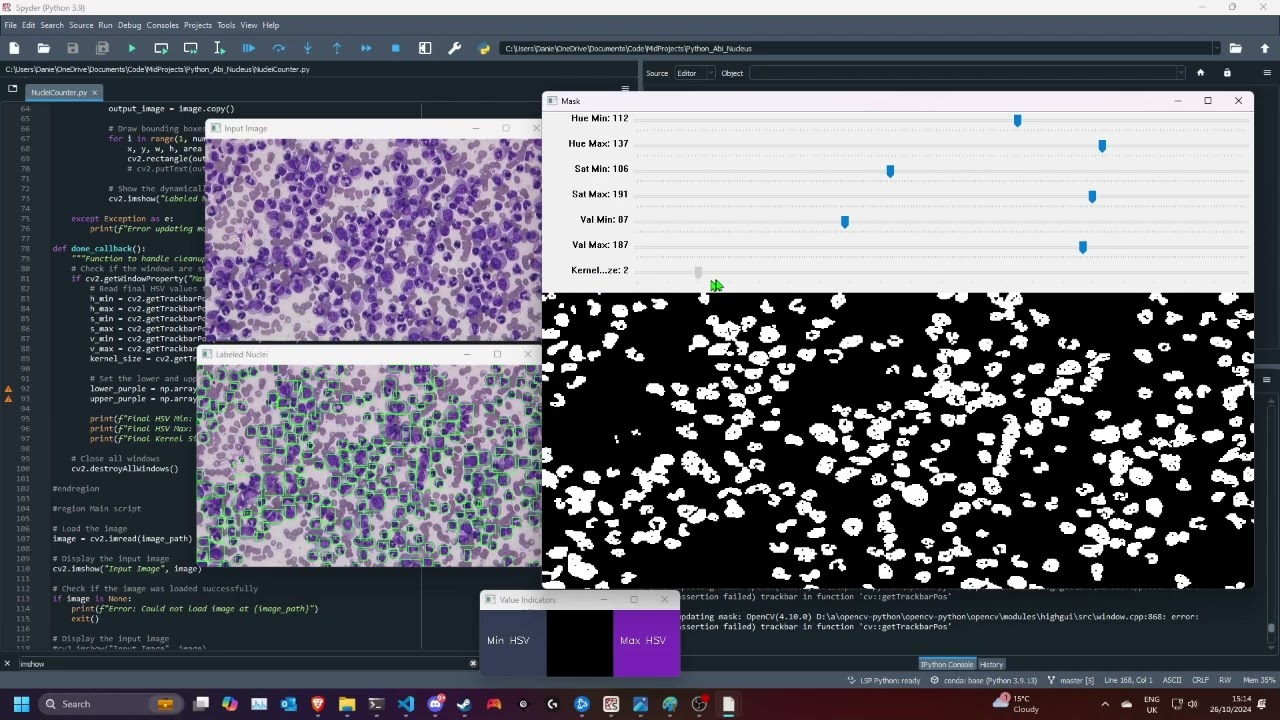
This Python script utilizes OpenCV to detect and label nuclei in an image based on user-defined HSV (Hue, Saturation, Value) color ranges. It allows for real-time adjustments of these ranges through trackbars and can filter out small detected nuclei based on area.
- Dynamic HSV Thresholding: Users can adjust the HSV thresholds interactively using trackbars to fine-tune the detection of purple nuclei in the image.
- Mask Cleaning Option: The script can clean the mask using morphological operations to reduce noise, with a toggle to include or exclude small areas from the analysis.
- Nuclei Detection: It employs connected components analysis to identify and label each detected nucleus in the binary mask.
- Output: The script generates an output image with bounding boxes around detected nuclei and saves it to disk.
- Python 3.x
- OpenCV (
cv2) - NumPy
To run the script, ensure you have your environment set up with the required packages, and execute:
python NucleiCounter.py-
You will be prompted to enter the: Image Path
-
You can either use the default values by just pressing
Enteror input your own values.
-
A window will pop up with:
- The original image
- The Labelled Nuclei
- A mask selector, which will allow you to select the regions of the image that contain the nuclei, as determined by its HSV values (and a kernel size = degree of cleaning)
- Value Indicators indicating the max and min hsv values
-
Use this to adjust the mask to only include the nuclei, use the value indicators to hepl you select a valid colour range, then compare the mask and labelled nuclei against the input image
-
Then when the mask looks good, press
d(for done) to continue.
- In the console you will now see the HSV Min, HSV Max, and Kernel Size values you selected (for reference) alongside the number of nuclei detected.
- The output image will be saved in the same directory as the script with the same name as the input image but with
_labeledappended to the name.