-
Notifications
You must be signed in to change notification settings - Fork 302
[ARCHIVE] caMicroscope Docker Distribution Guide
This page is severely out of date. Please see https://github.com/camicroscope/distro
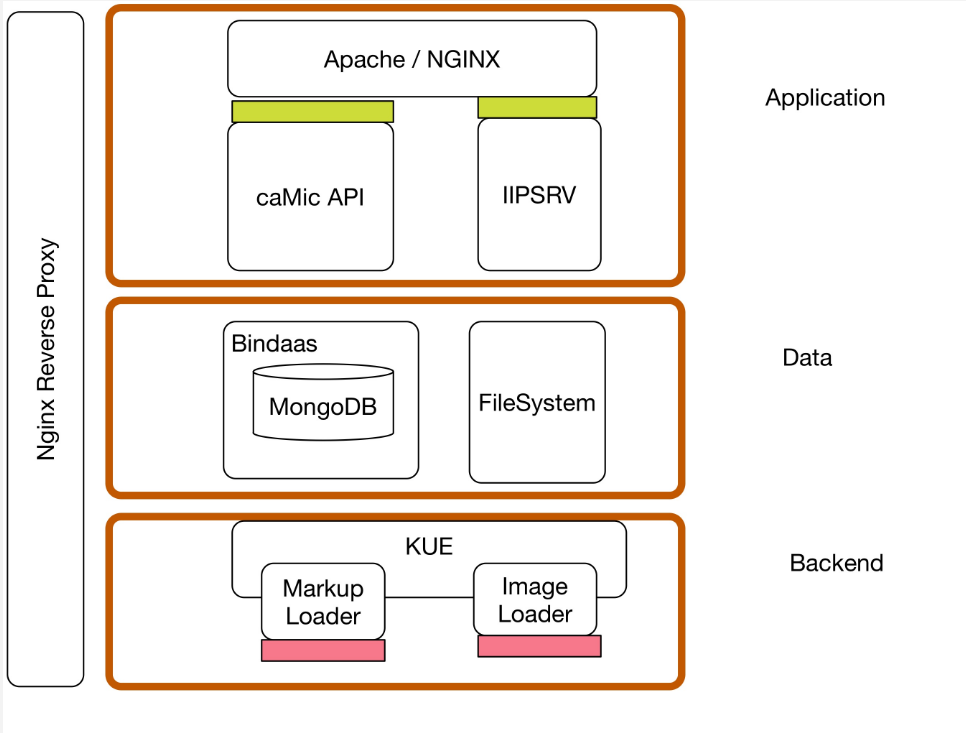
The goal of this guide is to be able to install all of caMicroscope componenets using docker containers. This guide will also guide you how to upload an image, its markups and see them in caMicroscope viewer.
Please make sure you have docker and git installed.
git clone https://github.com/camicroscope/DataDockerContainergit clone https://github.com/camicroscope/LoaderDockerContainergit clone https://github.com/camicroscope/ViewerDockerContainer
docker build -t camicroscope_data DataDockerContainerdocker build -t camicroscope_loader LoaderDockerContainerdocker build -t camicroscope_viewer ViewerDockerContainer
-
./install.shwill run the 3 containers in default configuration
Edit /var/www/html/camicroscope2/api/Configuration/config.php to set the $baseUrl to the IP of dataDockerContainer.
Lets assume you have an image TCGA-A1-A0SD-01A-01-BS1.svs on your file system.
To view this image in caMicroscope you'll need to load the image through caMicroscope data loader.
curl -v -F case_id=TCGA-A1-A0SD -F [email protected] http://localhost:6002/submitData
or go to
http://localhost:6002 in your browser to use the uploader UI.
You should get a success message.
Open http://localhost:1337/camicroscope2/osdCamicroscope.php?tissueId=TCGA-A1-A0SD to view the image. Please note the tissueId parameter is the case_id you supplied while loading the image