DIRAC is a Python package, written in PyTorch and based on Scanpy.
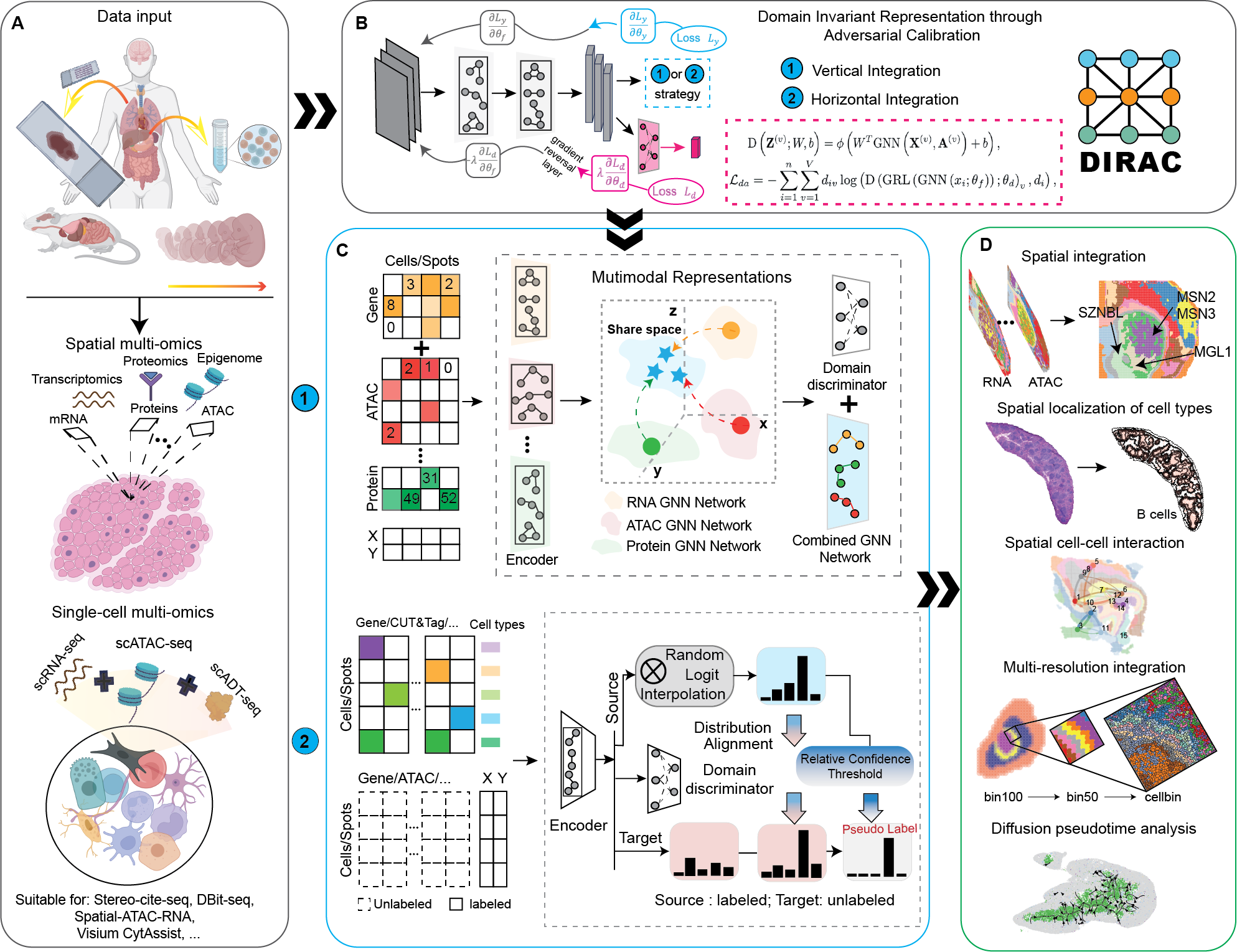
DIRAC is a graph neural network to integrate spatial multi-omic data into a unified domain-invariant embedding space and to automate cell-type annotation by transferring labels from reference spatial or single-cell multi-omic data.
DIRAC primarily includes two integration paradigms: vertical integration and horizontal integration, which differ in their selection of anchors. In vertical integration, multiple data modalities from the same cells are jointly analyzed, using cell correspondences in single-cell data or spot correspondences in spatial data as anchors for alignment. In horizontal integration, the same data modality from distinct groups of cells is aligned using genomic features as anchors. The best way to familiarize yourself with DIRAC is to check out our tutorial and our documentation.
For more details, please check out our publication.
.
├── sodirac # Main Python package
├── data # Data files
├── docs # Documentation files
├── environment.yaml # Reproducible Python environment via conda
├── requirements.yaml # Python packages required for issuing DIRAC
├── LICENSE
└── README.md
To install DIRAC, make sure you have PyTorch and PyG installed. For more details on dependencies, refer to the environment.yml file.
conda create -n dirac-env python=3.9 r-base=4.3.1 rpy2 r-mclust r-yarrr
Activate the environment and install PyTorch and PyG. Adjust the installation commands based on your CUDA version or choose the CPU version if necessary.
- General Installation Command
conda activate dirac-env
pip install torch==2.1.0 torchvision==0.16.0 torchaudio==2.1.0 --index-url https://download.pytorch.org/whl/cu118
pip install pyg_lib==0.3.1+pt21cu118 torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.1.0+cu118.html
pip install torch_geometric==2.3.1
- Tips for selecting the correct CUDA version
- Run the following command to verify CUDA version:
nvcc --version- Alternatively, use:
nvidia-smi - Modify installation commands based on CUDA
- For CUDA 12.1
pip install torch==2.1.0 torchvision==0.16.0 torchaudio==2.1.0 --index-url https://download.pytorch.org/whl/cu121 pip install pyg_lib==0.3.1+pt21cu121 torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.1.0+cu121.html pip install torch_geometric==2.3.1 - For CPU-only
pip install torch==2.1.0 torchvision==0.16.0 torchaudio==2.1.0 --index-url https://download.pytorch.org/whl/cpu pip install pyg_lib==0.3.1+pt21cpu torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.1.0+cpu.html pip install torch_geometric==2.3.1
- For CUDA 12.1
pip install sodirac
import sodirac as sd
Installing within a conda environment is recommended.
Please checkout the documentations and tutorials at dirac.readthedocs.io.