EnrichGT <- Fast, light weight enrichment analysis + insightful re-clustering results make all results explainable + Pretty HTML tables, Just in ONE package, designed for researchers in wet-labs.
Please see the package website for more info: https://zhimingye.github.io/EnrichGT/
-
Efficient C++-based functions for rapid enrichment analysis
-
Simple input format, empowering non-pro users
-
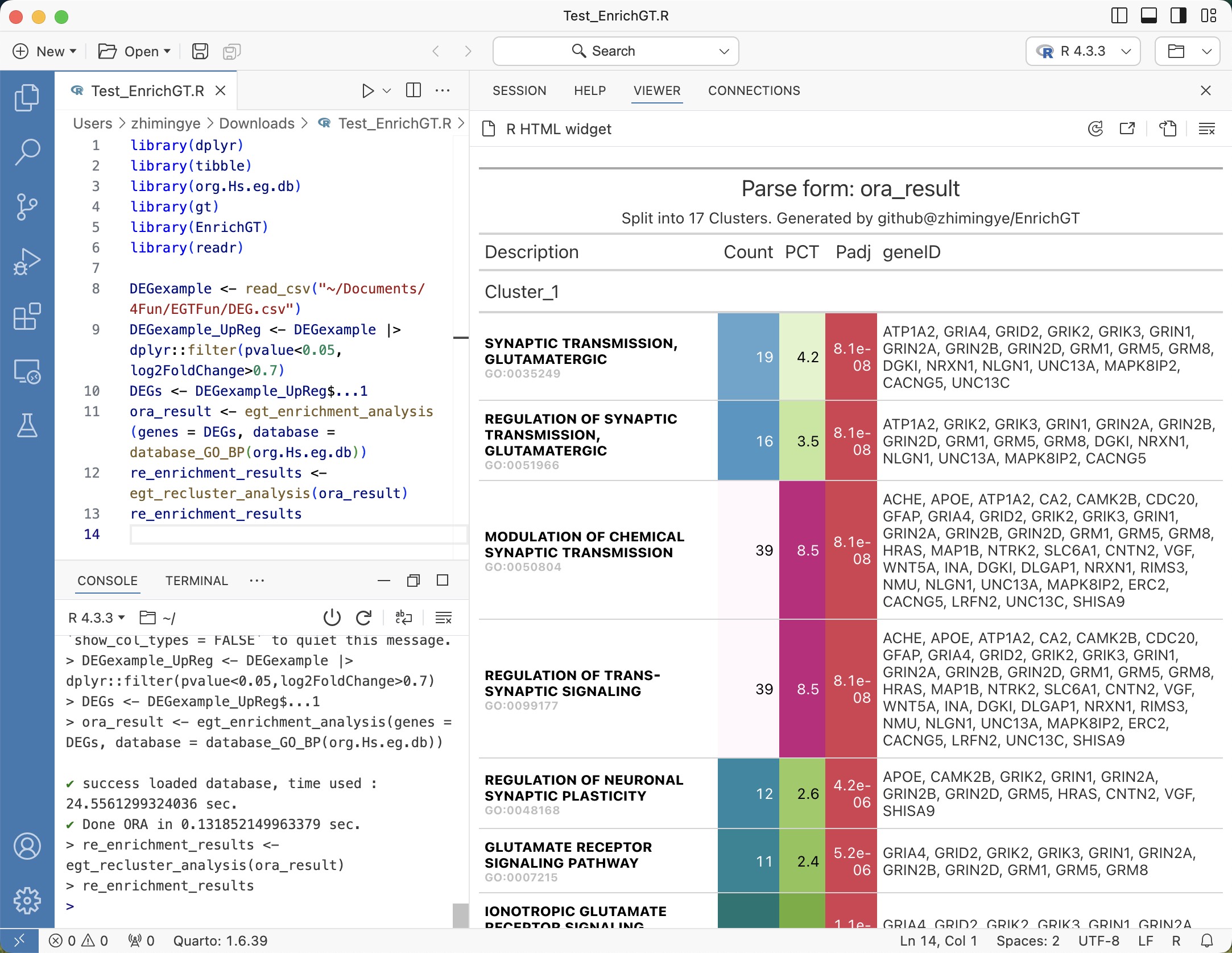
Re-clustering of enriched results provides clear and actionable insights
-
User-friendly HTML output that is easy to read and interpret
-
Do a series of things just in ONE package
install.packages("pak")
pak::pkg_install("ZhimingYe/EnrichGT")graph LR
M[genes]
N[genes with weights]
subgraph Enrichment Analysis
A[egt_enrichment_analysis]
B[egt_gsea_analysis]
end
subgraph Pathway Databases
D[database_* funcs]
end
subgraph Visualize results
P1[egt_plot_results]
P2[egt_plot_umap]
end
subgraph egt_recluster_analysis
K1[Pretty table]
CC[cluster modules]
MG[gene modules]
end
subgraph Pathway Act. and TF infer
I[egt_infer]
end
M --> A
N --> B
D --> A
D --> B
A --> C[Enriched Result]
B --> C
C --> CC
C --> MG
C --> P1
CC --> K1
MG --> K1
CC --> P1
CC --> P2
MG --> I