#Spatial Data in R ##Sydney Institute of Marine Science 10th August 2015
This work is published under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
![]()
##Hello World. Initial Steps
###Set up a project directory
###A fun intro
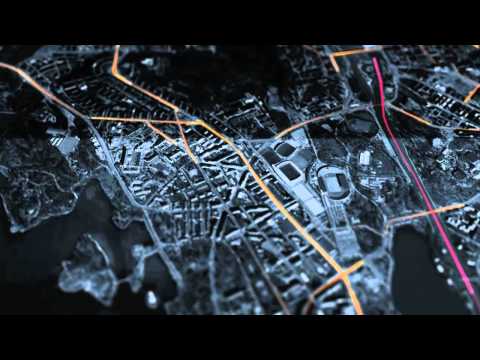
This exercise is from James Cheshire, a very cool analyst from University College London. His site is spatial.ly. His Population Lines image now hangs in my loungeroom!
This graphic uses ggplot (which we'll get to later...) to simply plot the routes that south England residents too and from work each day.
###Mapping Journeys to Work in the UK
library(plyr)
library(ggplot2)
library(maptools)
##read in the data- it is a really big dataset! It will take some time to read in
input<-read.table("Data/wu03ew_v1.csv", sep=",", header=T)
##select only the origin, destination and total kms from the data
input<- input[,1:3]
names(input)<- c("origin", "destination","total")
##download and input the population centroids from
centroids <- read.csv("Data/msoa_popweightedcentroids.csv")
or.xy <- merge(input, centroids, by.x="origin", by.y="Code")
names(or.xy) <- c("origin", "destination", "trips", "o_name", "oX", "oY")
dest.xy <- merge(or.xy, centroids, by.x="destination", by.y="Code")
names(dest.xy) <- c("origin", "destination", "trips", "o_name", "oX", "oY","d_name", "dX", "dY")
##plotting in ggplot2
xquiet <- scale_x_continuous("", breaks=NULL)
yquiet <-scale_y_continuous("", breaks=NULL)
quiet <-list(xquiet, yquiet)
ggplot(dest.xy[which(dest.xy$trips>10),], aes(oX, oY))+
geom_segment(aes(x=oX, y=oY,xend=dX, yend=dY, alpha=trips), col="white")+
scale_alpha_continuous(range = c(0.03, 0.3))+
theme(panel.background = element_rect(fill='black',colour='black'))+quiet+coord_equal()###Some preliminaries
#####Installing rgdal on a Mac The Easy Way In R run: install.packages(‘rgdal’,repos=”http://www.stats.ox.ac.uk/pub/RWin“)
The Hard Way If the easy way doesn't work. Download and install GDAL 1.8 Complete and PROJ framework v4.7.0-2 from: http://www.kyngchaos.com/software/frameworks%29
Download the latest version of rgdal from CRAN. Currently 0.7-1 Run Terminal.app, and in the same directory as rgdal_0.7-1.tar.gz run:
R CMD INSTALL --configure-args='--with-gdal-config=/Library/Frameworks/GDAL.framework/Programs/gdal-config --with-proj-include=/Library/Frameworks/PROJ.framework/Headers --with-proj-lib=/Library/Frameworks/PROJ.framework/unix/lib' rgdal_0.7-1.tar.gz
####Git and GitHub GitHub is a platform for hosting and collaborating on projects. You don’t have to worry about losing data on your hard drive or managing a project across multiple computers — sync from anywhere. Most importantly, GitHub is a collaborative and asynchronous workflow for building software better, together.
-
Sign Up here. https://github.com/join
-
To create a new repository -Click the icon next to your username, top-right. -Name your repository hello-world. -Write a short description. -Select Initialize this repository with a README.
-
Navigate to LucasCo/HASpatial. There you will find the the course materials that we will be working from.
-
Fork the repo into your own GitHub account.
###A brief primer on R classes
Remember that:
x <- sum(c(2, 5, 7, 1))
x## [1] 15
x is an object in R. It has a class, which represents the way the data in x is stored.
class(x)## [1] "numeric"
we can see that the class of x is numeric.
Classes determine what actions functions are going to have on the object. These functions, for example, include print, summary, plot etc. If no method is avialable for a specific class then it will try and default to known generic methods.
When R was first being created, there were no real functions. They were introduced in 1992 in a form called 'S3' classes, or 'old-style classes'.
Think back to the classic data set shipped with R, `mtcars'.
data(mtcars)
class(mtcars)## [1] "data.frame"
typeof(mtcars)## [1] "list"
names(mtcars)## [1] "mpg" "cyl" "disp" "hp" "drat" "wt" "qsec" "vs" "am" "gear" "carb"
mtcars is a data.frame that is stored as a list (this is why things like lapply will work on data.frames).
Old style classes are generally lists with attributes and not defined formally. In 1998 new style S4 style classes were introduced. The main definition between old and new style classes that that the new S4 classes have clear, formal, definitions that can specify the type and storage structure for each of the components, called slots.
An understanding of these classes is important, as most spatial data in R uses new style, S4 classes.
###R and spatial
Different spatial classes in R inherit their attributes from other classes.
library(sp)getClass("Spatial")#getClass("Spatial")
#Class "Spatial" [package "sp"]
#Slots:
#
#Name: bbox proj4string
#Class: matrix CRS#
#Known Subclasses:
#Class "SpatialPoints", directly
#Class "SpatialMultiPoints", directly
#Class "SpatialGrid", directly
#Class "SpatialLines", directly
#Class "SpatialPolygons", directly
#Class "SpatialPointsDataFrame", by class "SpatialPoints", distance 2
#Class "SpatialPixels", by class "SpatialPoints", distance 2
#Class "SpatialMultiPointsDataFrame", by class "SpatialMultiPoints", distance 2
#Class "SpatialGridDataFrame", by class "SpatialGrid", distance 2
#Class "SpatialLinesDataFrame", by class "SpatialLines", distance 2
#Class "SpatialPixelsDataFrame", by class "SpatialPoints", distance 3
#Class "SpatialPolygonsDataFrame", by class "SpatialPolygons", distance 2
m <- matrix(c(0, 0, 1, 1), ncol = 2, dimnames = list(NULL, c("min", "max")))
print(m)## min max
## [1,] 0 1
## [2,] 0 1
crs <- CRS(projargs = as.character(NA))
print(crs)## CRS arguments: NA
S <- Spatial(bbox = m, proj4string = crs)
S## An object of class "Spatial"
## Slot "bbox":
## min max
## [1,] 0 1
## [2,] 0 1
##
## Slot "proj4string":
## CRS arguments: NA
- We first creat a bounding box by making a 2 x 2 matrix.
- Create a CRS object (a specific object defined in the package
sp). Give it no arguments. - Create our first
Spatialobject, calling itS. We can see it has abboxand a CRS (called a proj4string).
####2. Spatial Points
Let's start using some real data!
The data below is from the OpenFlights project. OpenFlights is a tool that lets you map your flights around the world, search and filter them in all sorts of interesting ways, calculate statistics automatically, and share your flights and trips with friends and the entire world (if you wish). It's also the name of the open-source project to build the tool.
They have put their data (and code) on a public GitHub repo. Download and load into R directly.
library(RCurl)
URL <- "https://raw.githubusercontent.com/jpatokal/openflights/master/data/airports.dat"
x <- getURL(URL)
airports <- read.csv(textConnection(x), header = F)colnames(airports) <- c("ID", "name", "city", "country", "IATA_FAA", "ICAO", "lat", "lon", "altitude", "timezone", "DST",
"TimeZone")
head(airports)## ID name city country IATA_FAA
## 1 1 Goroka Goroka Papua New Guinea GKA
## 2 2 Madang Madang Papua New Guinea MAG
## 3 3 Mount Hagen Mount Hagen Papua New Guinea HGU
## 4 4 Nadzab Nadzab Papua New Guinea LAE
## 5 5 Port Moresby Jacksons Intl Port Moresby Papua New Guinea POM
## 6 6 Wewak Intl Wewak Papua New Guinea WWK
## ICAO lat lon altitude timezone DST TimeZone
## 1 AYGA -6.081689 145.3919 5282 10 U Pacific/Port_Moresby
## 2 AYMD -5.207083 145.7887 20 10 U Pacific/Port_Moresby
## 3 AYMH -5.826789 144.2959 5388 10 U Pacific/Port_Moresby
## 4 AYNZ -6.569828 146.7262 239 10 U Pacific/Port_Moresby
## 5 AYPY -9.443383 147.2200 146 10 U Pacific/Port_Moresby
## 6 AYWK -3.583828 143.6692 19 10 U Pacific/Port_Moresby
The only difference between the Spatial and SpatialPoints class is the addition of the coords slot.
air_coords <- cbind(airports$lon, airports$lat)
air_CRS <- CRS('+proj=longlat +ellps=WGS84')
airports_sp <- SpatialPoints(air_coords, air_CRS)
summary(airports_sp)####Methods
#####Bounding Box
Note that we did not specify a Bounding Box even though it is an integral component of the Spatial class. A bounding box is automatically assigned by taking the minimum and maximum latitude and longitude of the data.
bbox(airports_sp)## min max
## coords.x1 -179.877 179.95100
## coords.x2 -90.000 82.51778
#####Projection and datum
All spatial objects have a generic method proj4string. This sets the coordinate reference of the associated spatial data. Proj4 strings are a compact, but slightly quirky looking, way of specifying location references. Using the Proj4 syntax, the complete parameters to specify a CRS can be given. For example the PROJ4 string we usually use in Sydney is +proj=utm +zone=56 +south +ellps=GRS80 +units=m +no_defs. Here we essentially state that the CRS we are using is UTM (a projected CRS) in Zone 56. Most commonly we can start with an unprojected CRS (simply the longitute and latitude) using the CRS in the code above +proj=longlat +ellps=WGS84.
Proj4 strings can be assigned and re-assigned. NOTE that re-assigning a CRS does does NOT transform it. Simply saying the proj=utm will not convert the coordinates from long/lat into UTM.
proj4string(airports_sp) <- CRS(as.character(NA))
summary(airports_sp)## Object of class SpatialPoints
## Coordinates:
## min max
## coords.x1 -179.877 179.95100
## coords.x2 -90.000 82.51778
## Is projected: NA
## proj4string : [NA]
## Number of points: 8107
proj4string(airports_sp) <- air_CRS#####Subsetting Subsetting SpatialPoints can be accomplished in the same manner as most other subsetting in R - using logical vectors.
airports_oz <- which(airports$country=='Australia')
head(airports_oz)## [1] 1928 2047 2101 2194 2214 2313
oz_coords <- coordinates(airports_sp)[airports_oz,]
head(oz_coords)## coords.x1 coords.x2
## [1,] 151.488 -32.7033
## [2,] 149.611 -32.5625
## [3,] 148.755 -20.2760
## [4,] 150.832 -32.0372
## [5,] 151.342 -32.7875
## [6,] 128.307 -17.5453
plot(oz_coords)You can also use the subsetting methods directly on the SpatialPoints.
summary(airports_sp[airports_oz,])## Object of class SpatialPoints
## Coordinates:
## min max
## coords.x1 -153.01667 159.0770
## coords.x2 -42.83611 28.3835
## Is projected: FALSE
## proj4string : [+proj=longlat +ellps=WGS84]
## Number of points: 263
####3. SpatialPointsDataFrame
Often we need to associate attribute data to simple point locations. The SpatialPointsDataFrame is the container for this sort of data. You can see from the diagram above that the SpatialPointsDataFrame object extends the SpatialPoints class, and inherits the bbox, proj4string and coords objects.
getClass('SpatialPointsDataFrame')## Class "SpatialPointsDataFrame" [package "sp"]
##
## Slots:
##
## Name: data coords.nrs coords bbox proj4string
## Class: data.frame numeric matrix matrix CRS
##
## Extends:
## Class "SpatialPoints", directly
## Class "Spatial", by class "SpatialPoints", distance 2
## Class "SpatialPointsNULL", by class "SpatialPoints", distance 2
##
## Known Subclasses:
## Class "SpatialPixelsDataFrame", directly, with explicit coerce
So far we have downloaded a data.frame, where 2 columns were latitude and longitude, and constructed a simple SpatialPoints object from it.
A SpatialPointsDataFrame simply connects the SpatialPoints with the rest of the attribute data.
A key consideration here is that the row.names of both the matrix of coordinates and the data need to match.
airports_spdf <- SpatialPointsDataFrame(coords=air_coords, data=airports, proj4string=air_CRS)
summary(airports_spdf)## Object of class SpatialPointsDataFrame
## Coordinates:
## min max
## coords.x1 -179.877 179.95100
## coords.x2 -90.000 82.51778
## Is projected: FALSE
## proj4string : [+proj=longlat +ellps=WGS84]
## Number of points: 8107
## Data attributes:
## ID name city
## Min. : 1 North Sea : 16 London : 21
## 1st Qu.:2092 All Airports : 10 New York : 13
## Median :4257 Railway Station: 9 Hong Kong: 12
## Mean :4766 Train Station : 8 Berlin : 10
## 3rd Qu.:7508 Vilamendhoo : 6 Paris : 10
## Max. :9541 Bus : 5 Chicago : 9
## (Other) :8053 (Other) :8032
## country IATA_FAA ICAO lat
## United States:1697 :2227 \\N :1258 Min. :-90.000
## Canada : 435 BFT : 2 : 63 1st Qu.: 8.825
## Germany : 321 ZYA : 2 EDDB : 2 Median : 34.988
## Australia : 263 %u0 : 1 OIIE : 2 Mean : 26.818
## Russia : 249 04G : 1 RPVM : 2 3rd Qu.: 47.958
## France : 233 06A : 1 UATE : 2 Max. : 82.518
## (Other) :4909 (Other):5873 (Other):6778
## lon altitude timezone DST
## Min. :-179.877 Min. :-1266.0 Min. :-12.0000 A:2037
## 1st Qu.: -79.022 1st Qu.: 38.0 1st Qu.: -5.0000 E:1852
## Median : 5.292 Median : 272.0 Median : 1.0000 N:1364
## Mean : -3.922 Mean : 933.4 Mean : 0.1692 O: 214
## 3rd Qu.: 49.786 3rd Qu.: 1020.0 3rd Qu.: 4.0000 S: 391
## Max. : 179.951 Max. :14472.0 Max. : 13.0000 U:2195
## Z: 54
## TimeZone
## America/New_York : 628
## America/Chicago : 373
## Europe/Berlin : 319
## America/Anchorage : 258
## Europe/Paris : 232
## America/Los_Angeles: 226
## (Other) :6071
We can shorten this by simply joining a SpatialPoints objects with the data.
airports_spdf <- SpatialPointsDataFrame(airports_sp, airports)####4. Lines, Lines, Lines!
Lines are tricky. In the old S language they were simply represented by a series of numbers seperated by an NA that represented the end of the line. In R the most basic representation of a line is the Line object, essentially a matrix of 2-D coordinates. Many of these Line objects form the Lines slot of a Lines object. Other spatial data like a CRS and the bounding box are included in SpatialLines objects.
getClass('SpatialLines')## Class "SpatialLines" [package "sp"]
##
## Slots:
##
## Name: lines bbox proj4string
## Class: list matrix CRS
##
## Extends: "Spatial", "SpatialLinesNULL"
##
## Known Subclasses: "SpatialLinesDataFrame"
library('maps')
library('maptools')
oz <- map('world', 'Australia', plot=FALSE)
oz_crs <- CRS('+proj=longlat +ellps=WGS84')
oz <- map2SpatialLines(oz, proj4string=oz_crs)
str(oz, max.level=2)## Formal class 'SpatialLines' [package "sp"] with 3 slots
## ..@ lines :List of 43
## ..@ bbox : num [1:2, 1:2] 112.9 -54.7 159 -10.1
## .. ..- attr(*, "dimnames")=List of 2
## ..@ proj4string:Formal class 'CRS' [package "sp"] with 1 slot
A common way of accessing (and if you're game, manipulating) sp objects is via the list style functions sapply or lapply.
For example to find the length of the Lines slot, or how many Line objects it contains we can use the following
len <- sapply(slot(oz, 'lines'), function(x) length(slot(x, 'Lines')))
len## [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [36] 1 1 1 1 1 1 1 1
Lets plot it for funsies. See if you can do it. Hint: just use the plot() function.
plot(oz)Lets use the area around Auckland as an example of working with polygon data.
auc_crs <- CRS('+proj=longlat +ellps=WGS84')
auc_shore <- MapGen2SL('Data/auckland_mapgen.dat', auc_crs)
summary(auc_shore)## Object of class SpatialLines
## Coordinates:
## min max
## x 174.2 175.3
## y -37.5 -36.5
## Is projected: FALSE
## proj4string : [+proj=longlat +ellps=WGS84]
Lets have a closer look at the Auckland lines data and see if any of the lines join up. i.e Do the begining and end coordinates match.
First lets just check if all the lines objects have only one line in them.
nz_lines <- slot(auc_shore, 'lines')
sapply(nz_lines, function(x) length(slot(x, 'Lines')))## [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [36] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [71] 1 1 1 1 1 1 1 1 1 1
So all the lines objects in the Auckland dataset consists of only one line. That makes things easier. Let see how many have beginning and end coordinates that match. Don't worry if you don't follow this code.
This is a bit in-depth and you really wouldn't do this in real life, but lets construct some polygon data from our New Zealand lines data.
auc_islands <- sapply(nz_lines, function(x){
crds <- slot(slot(x, 'Lines')[[1]], 'coords')
identical(crds[1,], crds[nrow(crds),])
})
table(auc_islands)## auc_islands
## FALSE TRUE
## 16 64
So we have a bunch of lines that are essentially islands. These can be made into proper polygons.
First look at the structure of the hierarchical nature of polygons. They're much the same as SpatialLines.
getClass("Polygon")## Class "Polygon" [package "sp"]
##
## Slots:
##
## Name: labpt area hole ringDir coords
## Class: numeric numeric logical integer matrix
##
## Extends: "Line"
getClass("Polygons")## Class "Polygons" [package "sp"]
##
## Slots:
##
## Name: Polygons plotOrder labpt ID area
## Class: list integer numeric character numeric
getClass("SpatialPolygons")## Class "SpatialPolygons" [package "sp"]
##
## Slots:
##
## Name: polygons plotOrder bbox proj4string
## Class: list integer matrix CRS
##
## Extends: "Spatial", "SpatialPolygonsNULL"
##
## Known Subclasses: "SpatialPolygonsDataFrame"
If we take only those lines in the Auckland data set that are closed lines we get
auc_shore <- auc_shore[auc_islands]
list_of_lines <- slot(auc_shore, 'lines')
islands_sp <- SpatialPolygons(lapply(list_of_lines, function(x) {
Polygons(list(Polygon(slot(slot(x, "Lines")[[1]], "coords"))),
ID=slot(x, "ID"))
}), proj4string=CRS("+proj=longlat +ellps=WGS84"))
summary(islands_sp)## Object of class SpatialPolygons
## Coordinates:
## min max
## x 174.30297 175.22791
## y -37.43877 -36.50033
## Is projected: FALSE
## proj4string : [+proj=longlat +ellps=WGS84]
getClass('SpatialPolygons')## Class "SpatialPolygons" [package "sp"]
##
## Slots:
##
## Name: polygons plotOrder bbox proj4string
## Class: list integer matrix CRS
##
## Extends: "Spatial", "SpatialPolygonsNULL"
##
## Known Subclasses: "SpatialPolygonsDataFrame"
slot(islands_sp, "plotOrder")## [1] 45 54 37 28 38 27 12 11 59 53 5 25 26 46 7 55 17 34 30 16 6 43 14
## [24] 40 32 19 61 42 15 50 21 18 62 23 22 29 24 44 13 2 36 9 63 58 56 64
## [47] 52 39 51 1 8 3 4 20 47 35 41 48 60 31 49 57 10 33
The key points know for Polygon data (and indeed for most Spatial data in R) is:
- data: This holds the data.frame
- polygons: This holds the coordinates of the polygons
- plotOrder: The order that the coordinates should be drawn
- bbox: The coordinates of the bounding box (edges of the shape file)
- proj4string: A character string describing the projection system
##Visualising Spatial Data
####Let's start basic.
##basic intro
data(meuse)
str(meuse)## 'data.frame': 155 obs. of 14 variables:
## $ x : num 181072 181025 181165 181298 181307 ...
## $ y : num 333611 333558 333537 333484 333330 ...
## $ cadmium: num 11.7 8.6 6.5 2.6 2.8 3 3.2 2.8 2.4 1.6 ...
## $ copper : num 85 81 68 81 48 61 31 29 37 24 ...
## $ lead : num 299 277 199 116 117 137 132 150 133 80 ...
## $ zinc : num 1022 1141 640 257 269 ...
## $ elev : num 7.91 6.98 7.8 7.66 7.48 ...
## $ dist : num 0.00136 0.01222 0.10303 0.19009 0.27709 ...
## $ om : num 13.6 14 13 8 8.7 7.8 9.2 9.5 10.6 6.3 ...
## $ ffreq : Factor w/ 3 levels "1","2","3": 1 1 1 1 1 1 1 1 1 1 ...
## $ soil : Factor w/ 3 levels "1","2","3": 1 1 1 2 2 2 2 1 1 2 ...
## $ lime : Factor w/ 2 levels "0","1": 2 2 2 1 1 1 1 1 1 1 ...
## $ landuse: Factor w/ 15 levels "Aa","Ab","Ag",..: 4 4 4 11 4 11 4 2 2 15 ...
## $ dist.m : num 50 30 150 270 380 470 240 120 240 420 ...
coordinates(meuse) <- c('x', 'y')
str(meuse)## Formal class 'SpatialPointsDataFrame' [package "sp"] with 5 slots
## ..@ data :'data.frame': 155 obs. of 12 variables:
## .. ..$ cadmium: num [1:155] 11.7 8.6 6.5 2.6 2.8 3 3.2 2.8 2.4 1.6 ...
## .. ..$ copper : num [1:155] 85 81 68 81 48 61 31 29 37 24 ...
## .. ..$ lead : num [1:155] 299 277 199 116 117 137 132 150 133 80 ...
## .. ..$ zinc : num [1:155] 1022 1141 640 257 269 ...
## .. ..$ elev : num [1:155] 7.91 6.98 7.8 7.66 7.48 ...
## .. ..$ dist : num [1:155] 0.00136 0.01222 0.10303 0.19009 0.27709 ...
## .. ..$ om : num [1:155] 13.6 14 13 8 8.7 7.8 9.2 9.5 10.6 6.3 ...
## .. ..$ ffreq : Factor w/ 3 levels "1","2","3": 1 1 1 1 1 1 1 1 1 1 ...
## .. ..$ soil : Factor w/ 3 levels "1","2","3": 1 1 1 2 2 2 2 1 1 2 ...
## .. ..$ lime : Factor w/ 2 levels "0","1": 2 2 2 1 1 1 1 1 1 1 ...
## .. ..$ landuse: Factor w/ 15 levels "Aa","Ab","Ag",..: 4 4 4 11 4 11 4 2 2 15 ...
## .. ..$ dist.m : num [1:155] 50 30 150 270 380 470 240 120 240 420 ...
## ..@ coords.nrs : int [1:2] 1 2
## ..@ coords : num [1:155, 1:2] 181072 181025 181165 181298 181307 ...
## .. ..- attr(*, "dimnames")=List of 2
## .. .. ..$ : chr [1:155] "1" "2" "3" "4" ...
## .. .. ..$ : chr [1:2] "x" "y"
## ..@ bbox : num [1:2, 1:2] 178605 329714 181390 333611
## .. ..- attr(*, "dimnames")=List of 2
## .. .. ..$ : chr [1:2] "x" "y"
## .. .. ..$ : chr [1:2] "min" "max"
## ..@ proj4string:Formal class 'CRS' [package "sp"] with 1 slot
## .. .. ..@ projargs: chr NA
plot(meuse)data(meuse.riv)
Polygon(meuse.riv)## An object of class "Polygon"
## Slot "labpt":
## [1] 180182.6 331122.5
##
## Slot "area":
## [1] 2122714
##
## Slot "hole":
## [1] FALSE
##
## Slot "ringDir":
## [1] 1
##
## Slot "coords":
## [,1] [,2]
## [1,] 182003.7 337678.6
## [2,] 182136.6 337569.6
## [3,] 182252.1 337413.6
## [4,] 182314.5 337284.7
## [5,] 182331.5 337122.3
## [6,] 182323.9 336986.2
## [7,] 182268.2 336832.6
## [8,] 182160.9 336655.2
## [9,] 181979.1 336429.5
## [10,] 181831.8 336190.0
## [11,] 181739.6 335986.8
## [12,] 181675.3 335763.7
## [13,] 181658.4 335634.2
## [14,] 181656.0 335462.9
## [15,] 181675.8 335298.4
## [16,] 181719.8 334886.7
## [17,] 181718.5 334648.8
## [18,] 181679.7 334431.2
## [19,] 181610.3 334246.2
## [20,] 181451.8 334036.9
## [21,] 181325.1 333902.8
## [22,] 181029.8 333659.8
## [23,] 180915.2 333552.3
## [24,] 180740.8 333269.0
## [25,] 180579.2 332791.5
## [26,] 180528.2 332688.4
## [27,] 180401.2 332495.6
## [28,] 180309.3 332403.3
## [29,] 180208.2 332320.8
## [30,] 179871.9 332233.2
## [31,] 179752.2 332167.2
## [32,] 179569.6 332020.7
## [33,] 179508.4 331895.7
## [34,] 179420.9 331549.2
## [35,] 179286.5 331321.7
## [36,] 179033.3 331056.8
## [37,] 178895.9 330931.0
## [38,] 178708.0 330721.2
## [39,] 178551.1 330484.9
## [40,] 178474.2 330268.2
## [41,] 178467.9 330173.2
## [42,] 178499.5 329972.1
## [43,] 178547.1 329872.1
## [44,] 178641.8 329757.9
## [45,] 178725.5 329694.9
## [46,] 178819.0 329641.1
## [47,] 178922.3 329613.7
## [48,] 179059.1 329601.6
## [49,] 179212.0 329615.7
## [50,] 179343.8 329665.4
## [51,] 179447.2 329719.2
## [52,] 179535.0 329783.1
## [53,] 179596.4 329852.6
## [54,] 179834.7 330110.0
## [55,] 179891.5 330197.1
## [56,] 180043.6 330284.2
## [57,] 180187.0 330301.9
## [58,] 180279.0 330298.9
## [59,] 180404.6 330253.6
## [60,] 180713.7 330019.7
## [61,] 180799.1 329941.7
## [62,] 180947.4 329805.2
## [63,] 180982.3 329755.4
## [64,] 181065.6 329581.2
## [65,] 181114.0 329411.3
## [66,] 181124.0 329230.0
## [67,] 181098.1 329116.5
## [68,] 181008.6 329001.9
## [69,] 180859.0 328892.4
## [70,] 180653.9 328835.6
## [71,] 180493.1 328774.1
## [72,] 180314.2 328643.3
## [73,] 180263.8 328559.2
## [74,] 180186.3 328323.5
## [75,] 180167.3 328130.4
## [76,] 180120.6 327963.6
## [77,] 179890.0 327316.9
## [78,] 179803.4 327192.7
## [79,] 179546.9 326921.5
## [80,] 179397.3 326815.2
## [81,] 179233.7 326566.4
## [82,] 179102.6 326345.2
## [83,] 178983.9 326114.0
## [84,] 178916.6 325897.1
## [85,] 178868.8 325698.5
## [86,] 178766.0 325759.0
## [87,] 178873.6 326139.8
## [88,] 178983.0 326377.6
## [89,] 179137.2 326629.8
## [90,] 179282.3 326818.2
## [91,] 179556.9 327130.8
## [92,] 179651.5 327210.2
## [93,] 179823.9 327473.8
## [94,] 180000.0 327954.8
## [95,] 180064.8 328200.4
## [96,] 180087.1 328495.0
## [97,] 180111.8 328573.6
## [98,] 180178.4 328666.7
## [99,] 180264.0 328759.2
## [100,] 180448.6 328870.8
## [101,] 180812.8 329033.7
## [102,] 180903.7 329094.2
## [103,] 180970.5 329196.9
## [104,] 180977.8 329323.7
## [105,] 180965.9 329444.7
## [106,] 180940.1 329531.3
## [107,] 180901.6 329618.3
## [108,] 180773.4 329788.8
## [109,] 180671.6 329855.8
## [110,] 180569.8 329922.8
## [111,] 180386.6 330089.0
## [112,] 180274.3 330153.0
## [113,] 180189.1 330168.4
## [114,] 180096.3 330149.2
## [115,] 180000.0 330089.4
## [116,] 179916.9 330002.5
## [117,] 179836.6 329878.1
## [118,] 179691.9 329724.0
## [119,] 179617.3 329665.7
## [120,] 179512.7 329583.7
## [121,] 179307.2 329514.1
## [122,] 179239.9 329507.2
## [123,] 179058.6 329491.5
## [124,] 178843.6 329516.4
## [125,] 178649.3 329598.9
## [126,] 178519.4 329704.7
## [127,] 178400.1 329845.1
## [128,] 178328.1 329974.5
## [129,] 178304.0 330111.8
## [130,] 178315.0 330257.5
## [131,] 178354.5 330396.0
## [132,] 178413.6 330552.8
## [133,] 178643.8 330850.5
## [134,] 178840.0 331069.4
## [135,] 178987.6 331213.9
## [136,] 179129.8 331387.1
## [137,] 179208.3 331457.6
## [138,] 179299.2 331619.8
## [139,] 179334.0 331707.6
## [140,] 179375.0 331896.8
## [141,] 179423.7 332025.4
## [142,] 179484.2 332116.0
## [143,] 179500.5 332140.5
## [144,] 179601.6 332222.9
## [145,] 179789.1 332324.8
## [146,] 179901.7 332357.7
## [147,] 179965.5 332376.3
## [148,] 180153.4 332490.9
## [149,] 180296.9 332606.9
## [150,] 180390.0 332737.3
## [151,] 180481.4 332909.0
## [152,] 180695.3 333432.4
## [153,] 180871.1 333661.7
## [154,] 181395.9 334130.0
## [155,] 181423.6 334170.2
## [156,] 181567.0 334377.7
## [157,] 181606.5 334519.2
## [158,] 181622.3 334715.3
## [159,] 181570.5 335323.9
## [160,] 181569.0 335519.7
## [161,] 181581.5 335709.6
## [162,] 181626.9 335936.5
## [163,] 181661.9 336024.3
## [164,] 181716.7 336162.0
## [165,] 181855.3 336427.2
## [166,] 182167.2 336848.5
## [167,] 182222.0 336973.6
## [168,] 182234.5 337060.8
## [169,] 182242.4 337115.7
## [170,] 182224.0 337230.5
## [171,] 182197.6 337298.0
## [172,] 182164.0 337354.6
## [173,] 182107.3 337450.0
## [174,] 181981.7 337587.3
## [175,] 181810.4 337684.8
## [176,] 182003.7 337678.6
meuse_lst <-list(Polygons(list(Polygon(meuse.riv)), "meuse.riv"))
meuse_sp <- SpatialPolygons(meuse_lst)
plot(meuse_sp, axes=F)
plot(meuse, add=T, pch=16, cex=0.5) Try adding axes by using the
Try adding axes by using the axes=T argument.
Note that R has reserved the space that would have been taken up by the axes, even though they aren't plotted. Using the par command we can change many graphical parameters. It's good practice to save the default par arguments.
##par arguements
oldpar = par(no.readonly = TRUE)
layout(matrix(c(1,2),1,2))
plot(meuse, axes = TRUE, cex = 0.6)
title("Sample locations")
par(mar=c(0,0,0,0)+.1)
plot(meuse, axes = FALSE, cex = 0.6)
box()
par(oldpar)##Excercises
- In the data folder you'll find a list of all airports in the world. Read in that data, subset to all airports in Canada. Create a SpatialPointsDataFrame of this data and create a nice plot of all Canadian airports. note In the Data folder you will find a Shapefile of the Canadian coastline. We haven't covered this yet, but you can input, and plot this shapefile using the following code:
install.packages('rgdal')
library(rgdal) ##we will cover this in the following section
can_border <- readOGR(dsn='Data', layer='Canada')
can_border <- spTransform(can_border, CRS('+proj=longlat +ellps=WGS84'))
plot(can_border)####rgdal and importing other spatial data types
The GDAL (Geospatial Data Abstraction Library) is a library for reading and writing raster geospatial data formats, and is released under the permissive X/MIT style free software license by the Open Source Geospatial Foundation. As a library, it presents a single abstract data model to the calling application for all supported formats. It may also be built with a variety of useful command-line utilities for data translation and processing.
Calls to the GDAL framework are made using the r package rgdal. You should now have this on your computer. Hopefully it didn't cause too much trouble!
The main reason this package is so important for working scientists and analysts, is that it allows the import and export of the common ESRI data, shapefiles.
install.packages('rgdal')
library('rgdal')Reef Life Survey
RLS consists of a network of trained, committed recreational SCUBA divers, and an Advisory Committee made up of managers and scientists with direct needs for the data collected, and recreational diver representatives.
The RLS diver network undertakes scientific assessment of reef habitats using visual census methods, through a combination of targeted survey expeditions organised at priority locations under the direction of the Advisory Committee, and through the regular survey diving activity of trained divers in their local waters.
Here is a subset of there data available from the Integrated Marine Observing System, accessed here https://imos.aodn.org.au/imos123/home.
Lets read it in and then map their Sydney sites. I have exported raw data and converted to an ESRI Shapefile for use in most GIS.
The key component of reading in Shapefile data is to use the readOGR function. Note that several other methods exists, but the readOGR, from the rgdal package is most reliable and useful.
rls <- readOGR(dsn='Data', layer='rls_sites_sydney')## OGR data source with driver: ESRI Shapefile
## Source: "Data", layer: "rls_sites_sydney"
## with 133 features
## It has 6 fields
summary(rls)## Object of class SpatialPointsDataFrame
## Coordinates:
## min max
## coords.x1 325694.1 343135.4
## coords.x2 6228550.2 6260854.4
## Is projected: TRUE
## proj4string :
## [+proj=utm +zone=56 +south +ellps=WGS84 +units=m +no_defs]
## Number of points: 133
## Data attributes:
## Site SurveyID lat
## Grotto Point Lighthouse: 7 Min. : 2000854 Min. :-34.07
## Bare Island : 5 1st Qu.: 2001100 1st Qu.:-33.86
## Camp Cove NE : 5 Median : 2001289 Median :-33.84
## Fairlight : 5 Mean : 2602891 Mean :-33.86
## The Gap : 5 3rd Qu.: 2001926 3rd Qu.:-33.82
## Browns Rock : 4 Max. :10000059 Max. :-33.78
## (Other) :102
## lon richness totalfish
## Min. :151.1 Min. : 5.00 Min. : 30
## 1st Qu.:151.3 1st Qu.:16.00 1st Qu.: 367
## Median :151.3 Median :22.00 Median : 862
## Mean :151.3 Mean :21.48 Mean :1021
## 3rd Qu.:151.3 3rd Qu.:26.00 3rd Qu.:1316
## Max. :151.3 Max. :45.00 Max. :6502
##
plot(rls, pch=16)lets also bring in a Sydney Harbour shoreline layer, also as an ESRI Shapefile.
syd <- readOGR(dsn='Data', layer='SH_est_poly_utm')## OGR data source with driver: ESRI Shapefile
## Source: "Data", layer: "SH_est_poly_utm"
## with 1 features
## It has 1 fields
The Sydney shoreline files are simply in Longitude and Latitude (using the WGS84 representation of the Earth). Lets change this CRS to represent a local projection commonly used here in New South Wales. It is simply the Universal Transverse Mercator.
The Universal Transverse Mercator (UTM) conformal projection uses a 2-dimensional Cartesian coordinate system to give locations on the surface of the Earth. Like the traditional method of latitude and longitude, it is a horizontal position representation, i.e. it is used to identify locations on the Earth independently of vertical position. However, it differs from that method in several respects.
The UTM system divides the Earth between 80°S and 84°N latitude into 60 zones, each 6° of longitude in width. Zone 1 covers longitude 180° to 174° W; zone numbering increases eastward to zone 60 that covers longitude 174 to 180 East.
The spTransorm function within rgdal can project our lat/lon CRS into the correct UTM zone. Here in Sydney, the UTM zone is 56.
syd <- spTransform(syd, CRS('+proj=utm +zone=56 +south +ellps=WGS84 +units=m +no_defs'))note the rls data is already projected, and this projection is brought into R from the dbf file of the ESRI data suite.
Lets plot this out nicely.
plot(syd, axes=F, col='grey')
plot(rls, pch=16, col='dodgerblue',add=T)
title('Reef Life Survey Sites - Sydney Area', line=0.5)
##add scale bar and
#install.packages('GISTools')
#library(GISTools)
map.scale(xc= 325000, yc= 6250000, len=2000, ndivs=2, units='km')
north.arrow(xb= 325000, yb= 6258000, len=100)##Two common spatial manipulations
####Point in Polygon Operations
The figure produced above has points that fall outside the actual Harbour. We can do a Point in Polygon operation to remove these points.
syd_pol <- as(syd, "SpatialPolygons")
over(rls, syd_pol) #which sites are 'over' the polygon. Creates ## 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
## NA NA NA NA NA NA NA NA 1 1 NA NA NA 1 1 NA NA NA
## 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
## NA 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
## 1 1 1 1 1 1 1 1 1 1 NA NA 1 1 NA NA 1 1
## 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
## NA 1 NA NA NA NA NA NA NA NA NA NA NA NA NA 1 1 1
## 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
## 1 1 1 1 1 1 NA 1 1 NA NA 1 1 1 1 1 1 1
## 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108
## 1 1 1 1 1 1 1 1 1 NA NA NA NA 1 1 1 1 1
## 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126
## 1 1 1 1 1 1 NA NA NA NA NA NA NA NA NA 1 1 1
## 127 128 129 130 131 132 133
## 1 1 NA NA NA NA NA
!is.na(over(rls, as(syd, "SpatialPolygons")))## 1 2 3 4 5 6 7 8 9 10 11 12
## FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE FALSE FALSE
## 13 14 15 16 17 18 19 20 21 22 23 24
## FALSE TRUE TRUE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE
## 25 26 27 28 29 30 31 32 33 34 35 36
## TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## 37 38 39 40 41 42 43 44 45 46 47 48
## TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE
## 49 50 51 52 53 54 55 56 57 58 59 60
## TRUE TRUE FALSE FALSE TRUE TRUE FALSE TRUE FALSE FALSE FALSE FALSE
## 61 62 63 64 65 66 67 68 69 70 71 72
## FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE
## 73 74 75 76 77 78 79 80 81 82 83 84
## TRUE TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE FALSE FALSE TRUE
## 85 86 87 88 89 90 91 92 93 94 95 96
## TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## 97 98 99 100 101 102 103 104 105 106 107 108
## TRUE TRUE TRUE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE
## 109 110 111 112 113 114 115 116 117 118 119 120
## TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
## 121 122 123 124 125 126 127 128 129 130 131 132
## FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE
## 133
## FALSE
inside_harb <- is.na(over(rls, syd_pol))
rls_harb <- rls[!inside_harb, ]
summary(rls_harb)## Object of class SpatialPointsDataFrame
## Coordinates:
## min max
## coords.x1 332894.6 342417.8
## coords.x2 6251610.5 6258572.0
## Is projected: TRUE
## proj4string :
## [+proj=utm +zone=56 +south +ellps=WGS84 +units=m +no_defs]
## Number of points: 79
## Data attributes:
## Site SurveyID lat
## Camp Cove NE : 5 Min. : 2000866 Min. :-33.86
## Fairlight : 5 1st Qu.: 2001234 1st Qu.:-33.85
## Chowder Bay : 4 Median : 2001353 Median :-33.83
## Inside North Head : 4 Mean : 2406619 Mean :-33.83
## Middle Head North East: 4 3rd Qu.: 2001930 3rd Qu.:-33.82
## Middle Head Sth : 4 Max. :10000059 Max. :-33.80
## (Other) :53
## lon richness totalfish
## Min. :151.2 Min. : 5.00 Min. : 38.0
## 1st Qu.:151.3 1st Qu.:19.00 1st Qu.: 347.0
## Median :151.3 Median :23.00 Median : 764.0
## Mean :151.3 Mean :22.35 Mean : 996.2
## 3rd Qu.:151.3 3rd Qu.:27.00 3rd Qu.:1228.5
## Max. :151.3 Max. :43.00 Max. :6502.0
##
plot(syd, col='lightgrey')
plot(rls_harb, col='dodgerblue', add=T, pch=18)
title('Reef Life Survey Sites - Sydney Harbour only', line=0.5)
map.scale(xc= 325000, yc= 6250000, len=2000, ndivs=2, units='kms')
north.arrow(xb= 325000, yb= 6258000, len=100)####Spatial Polygon Dissolves
Sometimes there is a need to dissolve smaller polygons into larger ones. Think, for example, about having a Spatial Polygon dataset of Postal codes that you need to aggregate up into states. We'll work with a dataset that notes the participation in school sport across areas of the greater London metro area. Pretend there is a need to aggregate this shapefile simply into the area of greater London.
london_sport <- readOGR(dsn="Data", layer="london_sport")## OGR data source with driver: ESRI Shapefile
## Source: "Data", layer: "london_sport"
## with 33 features
## It has 4 fields
plot(london_sport)proj4string(london_sport) <- CRS("+init=epsg:27700")## Warning in `proj4string<-`(`*tmp*`, value = <S4 object of class structure("CRS", package = "sp")>): A new CRS was assigned to an object with an existing CRS:
## +proj=tmerc +lat_0=49 +lon_0=-2 +k=0.9996012717 +x_0=400000 +y_0=-100000 +ellps=airy +units=m +no_defs
## without reprojecting.
## For reprojection, use function spTransform in package rgdal
london_sport$london <- rep(1, length(london_sport))
london <- unionSpatialPolygons(london_sport, IDs=london_sport$london)
plot(london)###Exercise Two
- There are a bunch of cool spatial methods that can improve the efficiency of your work.
One of them is random spatial sampling. Say you need to randomly position sampling sites across an area. The most basic way of choosing these sites might make use of
spsample. Use the help file onspsampleto pick, and then map, 40 Fish traps to be deployed into Sydney Harbour every year. Your boss wants a single map, with colour coded symbols denoting which of the three years the fish traps will be deployed i.e. three colours on the map.
###Publication ready mapping
We wouldn't really use the base plotting function for anything other than basic maps. We tend to use the very powerful ggplot2 package by Hadley Wickham.
Lets work with the London Sport Participation dataset to start.
##To get the shapefiles into a
##format that can be plotted we have to use the fortify() function.
##he “polygons” slot contains the geometry of the polygons in the form
##of the XY coordinates used to draw the polygon outline. While ggplot is good,
##there is still no way for it to work out what is going on there. i.e. we need to convert
##this geometry into a `normal` data.frame.
sport.f <- fortify(london_sport, region = "ons_label")
head(sport.f)## long lat order hole piece id group
## 1 531026.9 181611.1 1 FALSE 1 00AA 00AA.1
## 2 531554.9 181659.3 2 FALSE 1 00AA 00AA.1
## 3 532135.6 182198.4 3 FALSE 1 00AA 00AA.1
## 4 532946.1 181894.8 4 FALSE 1 00AA 00AA.1
## 5 533410.7 182037.9 5 FALSE 1 00AA 00AA.1
## 6 533842.7 180793.6 6 FALSE 1 00AA 00AA.1
##need to merge back in the attribute data
sport.f <- merge(sport.f, london_sport@data, by.x = "id", by.y = "ons_label")
head(sport.f)## id long lat order hole piece group name
## 1 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 2 00AA 531554.9 181659.3 2 FALSE 1 00AA.1 City of London
## 3 00AA 532135.6 182198.4 3 FALSE 1 00AA.1 City of London
## 4 00AA 532946.1 181894.8 4 FALSE 1 00AA.1 City of London
## 5 00AA 533410.7 182037.9 5 FALSE 1 00AA.1 City of London
## 6 00AA 533842.7 180793.6 6 FALSE 1 00AA.1 City of London
## Partic_Per Pop_2001 london
## 1 9.1 7181 1
## 2 9.1 7181 1
## 3 9.1 7181 1
## 4 9.1 7181 1
## 5 9.1 7181 1
## 6 9.1 7181 1
Map <- ggplot(sport.f, aes(long, lat, group = group, fill = Partic_Per)) +
geom_polygon() +
coord_equal() +
labs(x = "Easting (m)", y = "Northing (m)", fill = "% Sport Partic.") +
ggtitle("London Sports Participation")
Map##and if we wanted black and white for publication
Map + scale_fill_gradient(low = "white", high = "black")Remember faceting? Well that can be done with maps as well using ggplot. We need to use a function called melt and reshape to get the data in the right form though.
###faceting maps - courtesy of James Cheshire and spatial.ly
library(reshape2)
london.data <- read.csv("Data/census-historic-population-borough.csv")
head(london.data)## Area.Code Area.Name Pop_1801 Pop_1811 Pop_1821 Pop_1831
## 1 00AA City of London 129000 121000 125000 123000
## 2 00AB Barking and Dagenham 3000 4000 5000 6000
## 3 00AC Barnet 8000 9000 11000 13000
## 4 00AD Bexley 5000 6000 7000 9000
## 5 00AE Brent 2000 2000 3000 3000
## 6 00AF Bromley 8000 9000 11000 12000
## Pop_1841 Pop_1851 Pop_1861 Pop_1871 Pop_1881 Pop_1891 Pop_1901 Pop_1911
## 1 124000 128000 112000 75000 51000 38000 27000 20000
## 2 7000 8000 8000 10000 13000 19000 27000 39000
## 3 14000 15000 20000 29000 41000 58000 76000 118000
## 4 11000 12000 15000 22000 29000 37000 54000 60000
## 5 5000 5000 6000 19000 31000 65000 120000 166000
## 6 14000 16000 22000 42000 62000 83000 100000 116000
## Pop_1921 Pop_1931 Pop_1939 Pop_1951 Pop_1961 Pop_1971 Pop_1981 Pop_1991
## 1 14000 11000 9000 5000 4767 4000 5864 4230
## 2 44000 138000 184000 189000 177092 161000 149786 140728
## 3 147000 231000 296000 320000 318373 307000 293436 284106
## 4 76000 95000 179000 205000 209893 217000 215233 211404
## 5 184000 251000 310000 311000 295893 281000 253275 227903
## 6 127000 165000 237000 268000 294440 305000 296539 282920
## Pop_2001
## 1 7181
## 2 163944
## 3 314565
## 4 218301
## 5 263466
## 6 295535
london.data.melt <- melt(london.data, id = c("Area.Code", "Area.Name"))
london.data.melt[1:50,]## Area.Code Area.Name variable value
## 1 00AA City of London Pop_1801 129000
## 2 00AB Barking and Dagenham Pop_1801 3000
## 3 00AC Barnet Pop_1801 8000
## 4 00AD Bexley Pop_1801 5000
## 5 00AE Brent Pop_1801 2000
## 6 00AF Bromley Pop_1801 8000
## 7 00AG Camden Pop_1801 100000
## 8 00AH Croydon Pop_1801 8000
## 9 00AJ Ealing Pop_1801 7000
## 10 00AK Enfield Pop_1801 11000
## 11 00AL Greenwich Pop_1801 35000
## 12 00AM Hackney Pop_1801 50000
## 13 00AN Hammersmith and Fulham Pop_1801 10000
## 14 00AP Haringey Pop_1801 6000
## 15 00AQ Harrow Pop_1801 4000
## 16 00AR Havering Pop_1801 6000
## 17 00AS Hillingdon Pop_1801 9000
## 18 00AT Hounslow Pop_1801 14000
## 19 00AU Islington Pop_1801 65000
## 20 00AW Kensington and Chelsea Pop_1801 18000
## 21 00AX Kingston upon Thames Pop_1801 5000
## 22 00AY Lambeth Pop_1801 33000
## 23 00AZ Lewisham Pop_1801 16000
## 24 00BA Merton Pop_1801 5000
## 25 00BB Newham Pop_1801 8000
## 26 00BC Redbridge Pop_1801 4000
## 27 00BD Richmond upon Thames Pop_1801 15000
## 28 00BE Southwark Pop_1801 116000
## 29 00BF Sutton Pop_1801 5000
## 30 00BG Tower Hamlets Pop_1801 144000
## 31 00BH Waltham Forest Pop_1801 8000
## 32 00BJ Wandsworth Pop_1801 13000
## 33 00BK Westminster Pop_1801 231000
## 34 UKI1 Inner London Pop_1801 939000
## 35 UKI2 Outer London Pop_1801 162000
## 36 H Greater London Pop_1801 1097000
## 37 00AA City of London Pop_1811 121000
## 38 00AB Barking and Dagenham Pop_1811 4000
## 39 00AC Barnet Pop_1811 9000
## 40 00AD Bexley Pop_1811 6000
## 41 00AE Brent Pop_1811 2000
## 42 00AF Bromley Pop_1811 9000
## 43 00AG Camden Pop_1811 128000
## 44 00AH Croydon Pop_1811 10000
## 45 00AJ Ealing Pop_1811 8000
## 46 00AK Enfield Pop_1811 13000
## 47 00AL Greenwich Pop_1811 46000
## 48 00AM Hackney Pop_1811 64000
## 49 00AN Hammersmith and Fulham Pop_1811 13000
## 50 00AP Haringey Pop_1811 7000
##remember what sport.f id was! How cool, now we can merge these data.
head(sport.f)## id long lat order hole piece group name
## 1 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 2 00AA 531554.9 181659.3 2 FALSE 1 00AA.1 City of London
## 3 00AA 532135.6 182198.4 3 FALSE 1 00AA.1 City of London
## 4 00AA 532946.1 181894.8 4 FALSE 1 00AA.1 City of London
## 5 00AA 533410.7 182037.9 5 FALSE 1 00AA.1 City of London
## 6 00AA 533842.7 180793.6 6 FALSE 1 00AA.1 City of London
## Partic_Per Pop_2001 london
## 1 9.1 7181 1
## 2 9.1 7181 1
## 3 9.1 7181 1
## 4 9.1 7181 1
## 5 9.1 7181 1
## 6 9.1 7181 1
plot.data <- merge(sport.f, london.data.melt, by.x = "id", by.y = "Area.Code")
head(plot.data)## id long lat order hole piece group name
## 1 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 2 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 3 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 4 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 5 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 6 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## Partic_Per Pop_2001 london Area.Name variable value
## 1 9.1 7181 1 City of London Pop_1901 27000
## 2 9.1 7181 1 City of London Pop_1831 123000
## 3 9.1 7181 1 City of London Pop_1821 125000
## 4 9.1 7181 1 City of London Pop_1971 4000
## 5 9.1 7181 1 City of London Pop_1811 121000
## 6 9.1 7181 1 City of London Pop_2001 7181
##order so plots are sensible i.e. in time order
plot.data <- plot.data[order(plot.data$order), ]
head(plot.data)## id long lat order hole piece group name
## 1 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 2 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 3 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 4 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 5 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## 6 00AA 531026.9 181611.1 1 FALSE 1 00AA.1 City of London
## Partic_Per Pop_2001 london Area.Name variable value
## 1 9.1 7181 1 City of London Pop_1901 27000
## 2 9.1 7181 1 City of London Pop_1831 123000
## 3 9.1 7181 1 City of London Pop_1821 125000
## 4 9.1 7181 1 City of London Pop_1971 4000
## 5 9.1 7181 1 City of London Pop_1811 121000
## 6 9.1 7181 1 City of London Pop_2001 7181
##now simply facet them in the usual ggplot way!
ggplot(data = plot.data, aes(x = long, y = lat, fill = value, group = group)) +
geom_polygon() + geom_path(colour = "grey", lwd = 0.1) + coord_equal() +
facet_wrap(~variable)Now go back to the start of the workbook, and try and make sense of the London Commute map code.
Exercises Three
- In the data folder there is a file called
cts_srr_04_2015_pt. This is the Automatic Identification System data for all ships entering Australian waters for the month of April this year. Also in the folder, you will find a rectangular shapefile that covers the south east coast of Australia. Subset the AIS data to the South East coast of Australia. Plot this data in a nice graph (preferably using GGPLOT2). See if you can find the outline of Australia somewhere on the internet (obviously as a shapefile) and plot this too.
Check if you can see the data - your working directory should be this file's location, and your data should be in Data/
file.exists("Data/Cairns_Mangroves_30m.tif")
file.exists("Data/SST_feb_2013.img")
file.exists("Data/SST_feb_mean.img")You should also have a folder Output/ for when we export results
Install the packages we're going to need - raster for raster objects, dismo is SDM, rgdal to read and write various spatial data
install.packages(c('raster', 'dismo','rgdal', 'gstat'))Check that you can load them
library(raster)
library(dismo)
library(rgdal)Now we're ready to go, but firstly, what is a raster? Well, simply, it is a grid of coordinates for which we can define a value at certain coordinate locations, and we display the corresponding grid elements according to those values. The raster data is essentially a matrix, but a raster is special in that we define what shape and how big each grid element is, and usually where the grid should sit in some known space (i.e. a geographic projected coordinate system).
Make a raster object, and query it
x <- raster(ncol = 10, nrow = 10) # let's make a small raster
nrow(x) # number of pixels
ncol(x) # number of pixels
ncell(x) # total number of pixels
plot(x) # doesn't plot because the raster is empty
hasValues(x) # can check whether your raster has data
values(x) <- 1 # give the raster a pixel value - in this case 1
plot(x) # entire raster has a pixel value of 1 Make a random number raster so we can see what's happening a little easier
values(x) <- runif(ncell(x)) # each pixel is assigned a random number
plot(x) # raster now has pixels with random numbers
values(x) <- runif(ncell(x))
plot(x)
x[1,1] # we can query rasters (and subset the maaxtrix values) using standard R indexing
x[1,]
x[,1]Use this to interactively query the raster - press esc to exit
click(x)What's special about a raster object?
str(x) # note the CRS and extent, plus plenty of other slots
crs(x) # check what coordinate system it is in, the default in the PROJ.4 format
xmax(x) # check extent
xmin(x)
ymax(x)
ymin(x)
extent(x) # easier to use extent
res(x) # resolution
xres(x) # just pixel width
yres(x) # just pixel height- make a raster with a smiley face
- extract some vector and matrix data from the raster
- subset the raster into a smaller chunk (tricker - see
?"raster-package")
Import the Cairns mangrove data and have a look at it
mangrove <- raster("Data/Cairns_Mangroves_30m.tif")
crs(mangrove) # get projection
plot(mangrove, col = topo.colors("2")) # note two pixel values, 0 (not mangrove) and 1 (mangrove)
NAvalue(mangrove) <- 0 # make a single binary dataset with mangroves having a raster value 1
plot(mangrove, col = "mediumseagreen")The legend is a little odd - we can change to a categorical legend by doign this - but we'll stick to the default continous bar generally so as to reduce clutter in the code
cols <- c("white","red")
plot(mangrove, col=cols, legend=F)
legend(x='bottomleft', legend=c("no mangrove", "mangrove"), fill=cols)Simple processing
agg.mangrove <- aggregate(mangrove, fact=10) # aggregate/resample cells (10 times bigger)
par(mfrow=c(2,2))
plot(mangrove, col = "mediumseagreen")
plot(agg.mangrove, col = "firebrick")
plot(agg.mangrove, col = "firebrick")
plot(mangrove, col = "mediumseagreen", add=TRUE) # add information to current plotCreate a simple buffer
buf.mangrove <- buffer(agg.mangrove, width=1000) # add a buffer
par(mfrow=c(1,1))
plot(buf.mangrove, col = "peachpuff")
plot(mangrove, col = "mediumseagreen", add = T) # note add= argumentConvert raster to point data, and then import point data as raster
pts.mangrove <- rasterToPoints(mangrove)
str(pts.mangrove)
par(mfrow=c(2,2))
plot(mangrove)
plot(rasterFromXYZ(pts.mangrove)) # huh?
NAvalue(mangrove) <- -999
pts.mangrove <- rasterToPoints(mangrove)
plot(rasterFromXYZ(pts.mangrove))
NAvalue(mangrove) <- 0 # set it back to 0
par(mfrow=c(1,1))Export your data - lets try the aggregated raster
KML(agg.mangrove, "Output/agg.mangrove.kml", overwrite = TRUE)
writeRaster(agg.mangrove, "Output/agg.mangrove.tif", format = "GTiff")Hang on, what about multiband rasters? The raster package handles them in the same way, just the nbands() attribute is >1 - think about an array instead of a matrix
multiband <- raster("Data/multiband.tif")
nbands(multiband)
nrow(multiband)
ncol(multiband)
ncell(multiband) What about making our own multiband raster?
for (i in 1:4) { assign(x=paste0("band",i), value=raster(ncol=10,nrow=10))}
values(band1) <- runif(100); values(band2) <- runif(100); values(band3) <- runif(100); values(band4) <- runif(100)
multiband.stack <- stack(list(band1,band2,band3,band4))
nlayers(multiband.stack)
plot(multiband.stack)Plotting an RGB image?
plotRGB(multiband.stack, r=1, g=2, b=3)
range(multiband.stack)
plotRGB(multiband.stack, r=1, g=2, b=3, scale=1) # let it know what the max value is for display
plotRGB(multiband.stack, r=3, g=2, b=1, scale=1)
plotRGB(multiband.stack, r=2, g=3, b=4, scale=1)Other handy processing functions
?crop
?merge
?trim
?interpolate
?reclassify
?rasterToPolygonsSome handy analysis functions
?zonal # zonal statistics
?focal # moving windows
?calc # raster calculator
?distance # distance calculations
?sampleRandom
?sampleRegular
?sampleStratifiedWe won't go into detail on coordinate and projection systems today, but very briefly, remembering CRS() objects from the earlier sessions
crs(mangrove)
proj4string(mangrove)
latlong <- "+init=epsg:4326"
CRS(latlong)
eastnorth <- "+init=epsg:3857"
CRS(eastnorth)
latlongs.mangrove <- rasterToPoints(mangrove, spatial=T)
latlongs.mangrove
projected.pts.mangrove <- spTransform(latlongs.mangrove, CRS(eastnorth))
projected.pts.mangrove- import the raster "Landsat_TIR.tif" - it's a TIR (thermal infrared) image from the Landsat 8 satellite captured over a cropping area
- suppose we modelled the TIR values via linear regression to calculate the real on ground temperature, and beta0 was 0.5 and beta1 was 0.1 (i.e. y = 0.1x + 0.5) - make a map of temperature (hint:
?calc, and you'll need to write a function) - give the plot a title and axis labels, and colours that make sense for temperature
- make a matching raster (in extent and number of pixels, for the easiest solution) with zone codes (for each pixel), then calulate the mean/sd temperature in those zones (hint:
?valuesand?zonal)
Now let's take a bit of a whirlwind tour of the types of analyses we can do, and hopefully discover a bit deeper understanding of raster analysis in R.
Load up some SST data - Feb 2013 for the globe (as an aside, check this link for more great ocean global data sets: http://oceancolor.gsfc.nasa.gov/cms/)
sst.feb <- raster("Data/SST_feb_2013.img")
plot(sst.feb)Crop it to the pacific so we can compare our mangrove data
pacific.extent <- extent(mangrove) + 80 # take advantage of the way R handles vector arithmatic!
pacific.extent # check it
sst.feb.crop <- crop(sst.feb, pacific.extent) # crop to the pacific
plot (sst.feb.crop)Load up the long term mean SST data for Feb
sst.feb.mn <- raster("Data/SST_feb_mean.img")
plot(sst.feb.mn)
sst.mn.crop <- crop(sst.feb.mn, pacific.extent)
plot (sst.mn.crop)Now let's make an SST anomoly map
sst.anomaly <- sst.feb.crop - sst.mn.crop # R + {raster} matrix arithmatic
plot (sst.anomaly) # plot the anomaly map
plot(sst.anomaly, col = topo.colors("100")) # different colours
plot(sst.anomaly, col = rev(heat.colors("100"))) # heat colours
contour(sst.anomaly, add = T) # add contoursQuery single values,
minValue(sst.anomaly) # coldest pixel
maxValue(sst.anomaly) # warmest pixel
plot(sst.anomaly==maxValue(sst.anomaly))or plots/stats for the entire image,
plot(sst.anomaly > 1)
hist(sst.anomaly, main = "February SST Anomaly - Pacific", xlab = "sst anomaly")or let's be a litle more tricky!
max.anom <- which.max(sst.anomaly)
max.xy <- xyFromCell(sst.anomaly, max.anom)
plot(sst.anomaly, col = rev(heat.colors("100")))
points(max.xy, pch=8, cex=2)Sampling points conditionally? Sure. We'll see a better wrapper for this further down though.
xy <- xyFromCell(sst.anomaly,sample(1:ncell(sst.anomaly), 20)) # 20 random points
points(xy)
extract(sst.feb, xy)
?getValues # check this out tooRe-capping writing back to disk
# writing rasters
writeRaster(sst.anomaly, "Output/sst.anomaly.tif", format = "GTiff")
KML(sst.anomaly, "Output/sst.anomaly.kml")
save(sst.anomaly, file="Output/sst.anomaly.feb.RData")
save(sst.feb.mn, file="Output/sst.feb.mn.RData") # check the file size, huh?What's going on with those last two {r, eval=FALSE} save() commands? Something else to understand about the way the {r, eval=FALSE} raster package handles raster files is that for larger rasters, the whole file is not stored in memory, rather it is just a pointer to the file. You can test whether or not it is
inMemory(sst.feb.mn) # R will only access file when needed.
inMemory(sst.anomaly) # it's in memory. We saw stack() earlier, and we can use it for multi-band imagery, but also to stack up different information sources. brick() works in the same way, except that it is designed for smaller objects, and a RasterBrick can only point to one file, opposed to a RasterStack, which can point to multiple files.
sst.stack <- stack(sst.mn.crop, sst.feb.crop, sst.anomaly)
plot(sst.stack)
nlayers(sst.stack)
plot(sst.stack,2)
names(sst.stack)[3] <- "SST_anomaly"Now let's look at quick example of what we can do with rasters in context of species distribution modelling and spatial modelling. First lets extract some random points - make sure you've run library(dismo)
rpoints.sst <- randomPoints(sst.stack,500) #?randomPoints for more options
plot(sst.stack,2)
points(rpoints.sst, pch = 16, cex = 0.7)
sst.samp <- extract(sst.stack, rpoints.sst) # extract values through stack this time
str(sst.samp)
sst.samp <- data.frame(sst.samp)
plot(sst.samp$SST_anomaly ~ sst.samp$SST_feb_2013)What if we had some real biological data at those points? Well, let's make some up, and then fit a model to it
sst.samp$shark.abund <- rpois(n=nrow(sst.samp), lambda=round(sst.samp$SST_feb_2013))
plot(sst.samp$shark.abund ~ sst.samp$SST_feb_2013)
shark.glm <- glm(shark.abund ~ SST_feb_2013 + SST_anomaly,
data=sst.samp, family="poisson")
summary(shark.glm)We would usually use predict() on a model fit object, and we can use it similarly for predicting out to raster data
predict(shark.glm, type="response")
shark.predict <- predict(sst.stack, shark.glm, type='response')
plot(shark.predict, col=rev(rainbow(n=10, start=0, end=0.3)), main="Shark abundance as a function of SST")What if we just had the physical data at some points, and wanted to make those into a geographically weighted SST map? We'll use library(gstat) to try an IDW (inverse distance weighted) interpolation
plot(sst.feb)
rpoints.sst.feb <- randomPoints(sst.feb, 500)
points(rpoints.sst.feb)
rpoints.sst.feb <- data.frame(rpoints.sst.feb, extract(sst.feb, rpoints.sst.feb))
names(rpoints.sst.feb)[3] = "SST"
# fit the idw model
sst.idw.fit <- gstat(id="SST", formula=SST~1, locations=~x+y, data=rpoints.sst.feb,
nmax=5, set=list(idp=1))
sst.idw <- interpolate(sst.feb, sst.idw.fit)
par(mfrow=c(1,2))
plot(sst.feb); plot(sst.idw) # ahh!
sst.idw <- mask(sst.idw, sst.feb)
plot(sst.feb); plot(sst.idw)Let's have a go at ordinary kriging (ignoring assuptions about Gaussian repsosne data)
# first some package specific projection stuff
coordinates(rpoints.sst.feb) <- ~x+y
proj4string(rpoints.sst.feb) <- proj4string(sst.feb)
# now the kriging
sst.vario <- variogram(object=SST~1, data=rpoints.sst.feb) # could log(y) is SST wasn't negative
sst.vario.fit <- fit.variogram(sst.vario, vgm(psill=0.3, model="Gau", range=100)) #?vgm
# (Exp)ponential, (Sph)rical, (Gau)ssian, (Mat)ern, (Spl)ine, (Lin)ear etc.
sst.ordkirg.fit <- gstat(id="SST", formula=SST~1, model=sst.vario.fit, data=rpoints.sst.feb)
sst.ordkrig <- interpolate(sst.feb, sst.ordkirg.fit)
sst.ordkrig <- mask(sst.ordkrig, sst.feb)
plot(sst.feb); plot(sst.ordkrig)- try generating some stats (values or plots) for SST anomaly for different regions, either across the globe or across Australia
- try come band math, or some conditional statements using multiple rasters or a RasterStack
- create another SDM scenario - either using downloaded data, or totally simulated data
- do some more interpolations, varying the number of points used, and see how that effects your interpollated product
- try and figure our co-kriging, or interpolation with added covariate data (hint: there's an example in
?interpolate)
Let's get some climate data using the raster package
par(mfrow=c(1,1))
rain <- getData('worldclim', var="prec", res=10, lon=5, lat=45) # this will d/l data to getwd()
plot(rain)
nlayers(rain)
rpoints.rain <- randomPoints(rain,500) # through the stack
plot(rain, 1) # plot january rainfall
points(rpoints.rain, pch=16, cex=0.7)
samp.rain <- extract(rain, rpoints.rain)
head(samp.rain)Get maps using dismo
install.packages("XML"); library(XML)
Aus <- gmap("Australia") # get google maps normal
plot(Aus)
AusSat <- gmap("Australia", type="satellite") # get google maps satellite image
plot(AusSat)Get simple maps
install.packages("maps"); library(maps)
map(database = "world", col = "grey")A couple other handy packages
library(rasterVis)
library(maptools)