Type: Master's Thesis
Author: Dingyi Lai
1st Examiner: Prof. Dr. Stefan Lessmann
2nd Examiner: Prof. Dr. Sonja Greven
Synthetic Dataset:
The 911 Emergency Calls Dataset for Montgomery County
The estimation of causal effects becomes increasingly complex when interventions exert diverse influences across quantiles. Addressing this challenge, we introduce a novel global framework that seamlessly integrates causal analysis with prediction algorithms. Despite remaining theoretical gaps, we propose a standardized approach to answer this research question. This involves defining causal mechanisms via directed acyclic graphs, elucidating theoretical assumptions, conducting placebo tests to identify causal effects, and estimating probabilistic causal effects within the global causal framework. Through comparative analysis utilizing synthetic and real-world datasets, we demonstrate the potential of this framework to estimate varying causal effects across quantiles over time. While promising, ongoing refinement is necessary to enhance the framework's consistency and robustness.
Keywords: Causal Inference \sep Time-varying Effect \sep Probabilistic Prediction
Python >= 3.6
-
Clone this repository
-
Create an virtual environment and activate it
python -m venv thesis-env
source thesis-env/bin/activate- Install requirements
pip install --upgrade pip
pip install -r requirements.txt- Path Variables
Set the PYTHONPATH env variable of the system. Append absolute paths of both the project root directory and the directory of the src/models/DeepProbCP/cocob_optimizer into the PYTHONPATH for the model DeepProbCP
For R scripts, make sure to set the working directory to the project root folder.
We have used two datasets for the experiments of this paper:
- Synthetic Dataset:
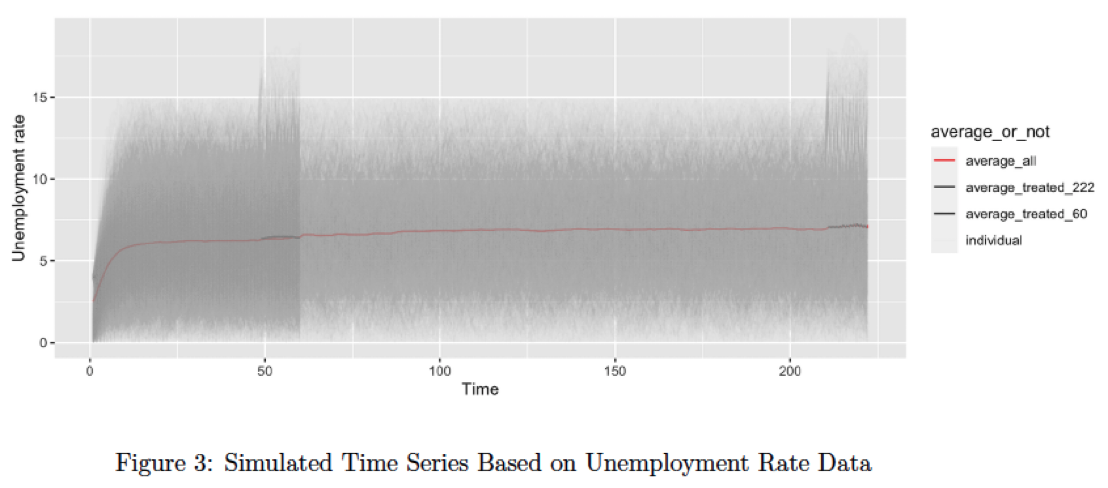
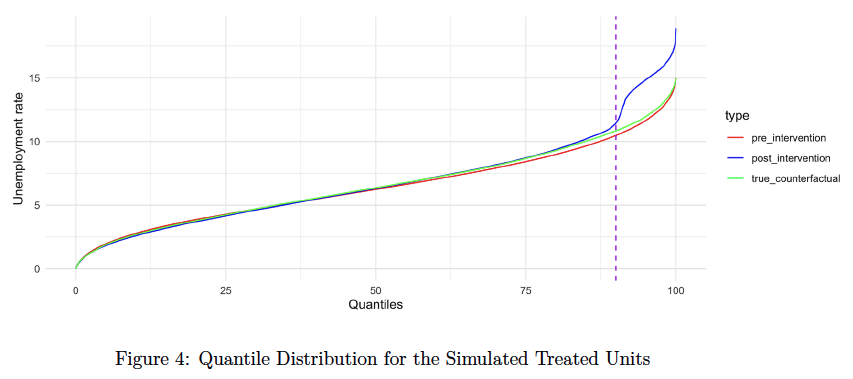
The synthetic dataset comprises 24 scenarios for generation, varying from short time series (with a length of 60) to long time series (with a length of 222). These scenarios range from homogeneous intervention over 0.9 quantiles (adding one unit standard deviation of treated units before intervention) to heterogeneous intervention over 0.9 quantiles (adding a random number between 0.7 to 1.5, multiplied by one unit standard deviation of treated units before intervention).
Furthermore, the dataset spans from a few time series (10 in total), medium time series (101 in total) to many time series (500 in total), and from a linear data generation process (autoregressive regression, i.e., AR) to a nonlinear structure (self-exciting threshold autoregressive, i.e., SETAR).
The codes for simulation and exploratory data analysis are located in src/prepare_source_data/sim/simulation.rmd, with its HTML version available at simulation.html. The original data for simulation is situated in ./data/text_data/sim/unrate.txt, accessible from Data_USEconModel in MATLAB.
For DeepProbCP, the dataset undergoes preprocessing using a window moving strategy, implemented in src/prepare_source_data/sim/preprocessing_layer.R. This aligns with the original codes from Grecov et al. for DeepCPNet. Subsequently, src/prepare_source_data/sim/create_tfrecords.py facilitates the creation of data in the tfrecords format to enhance computational efficiency, a method also employed in the previous codes for DeepCPNet. The customized module "tfrecords_handler" is also available in src/models/DeepProbCP.
- The 911 Emergency Calls Dataset for Montgomery County
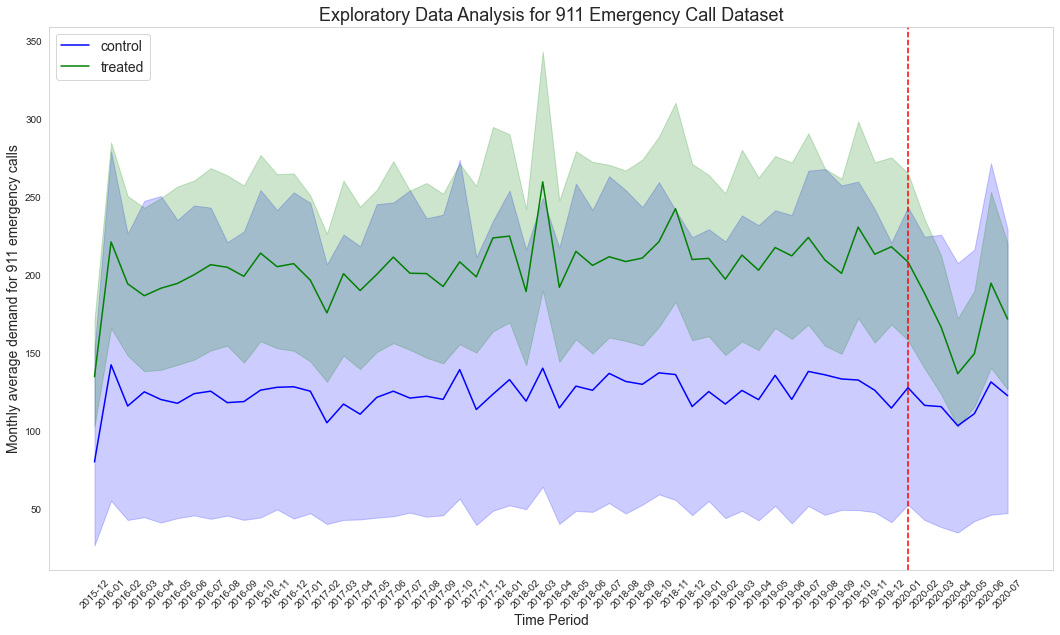
This dataset, sourced from Kaggle, contains records of 911 emergency calls from Montgomery County, Pennsylvania, USA, spanning from December 2015 to July 2020. The raw data file, named "911.csv," is available for download from the following https://www.kaggle.com/mchirico/montcoalert.
Exploratory Data Analysis:
- The exploratory data analysis is documented in
src/prepare_source_data/calls911/EDA.ipynb.
Data Wrangling:
- To process the data, the script
src/prepare_source_data/calls911/calls911_wrangling_code.Ris utilized for tasks such as splitting, formatting, and cleaning. - The resulting output data should ideally be stored in
./data/text_data/calls911/calls3.csv. However, due to size limitations on GitHub, this file has been removed from the current repository.
Preprocessing:
- Further preprocessing steps are conducted using
src/prepare_source_data/calls911/calls911_to_forecasting_code.R. - The full monthly dataset is available in
./data/text_data/calls911/calls911_month_full.txt, while the training dataset spans from December 2015 to December 2019 and is stored in./data/text_data/calls911/calls911_month_train.txt. The testing dataset spans from January 2020 to July 2020 and is stored in./data/text_data/calls911/calls911_month_test.txt. - To address the training and testing datasets in subsequent steps, scripts
src/prepare_source_data/calls911/adjustOrigTrainDtSet.Randsrc/prepare_source_data/calls911/adjustOrigTestDtSet.Rare utilized. The resulting output files are namedcallsMT2_train.csvandcallsMT2_test_actual.csv, serving as the training and testing datasets, respectively.
For DeepProbCP:
- The dataset undergoes preprocessing using a window moving strategy after being transformed to tfrecords by
src/prepare_source_data/calls911/create_tfrecords.py. - These preprocessing steps are implemented in
src/prepare_source_data/calls911/mean_stl_train_validation_withoutEXvar.Randsrc/prepare_source_data/calls911/mean_stl_test_withoutEXvar.R.
For Other Models:
- The script
src/prepare_source_data/calls911/benchmarks.pyis utilized to convertcalls911_month_full.txtto a standardized input format, resulting in./data/text_data/calls911/calls911_benchmarks.csv.
To train the model(s) in the paper, for
-
Causal Impact Run the codes in
src/models/CausalImpact/causalimpact.ipynb -
TSMixer Run the codes in
src/models/TSMixer/tsmixer.ipynb -
DeepProbCP Run the codes in
src/models/DeepProbCP/DeepProbCP.sh
The model expects a number of arguments.
- dataset_type - synthetic or real-world dataset
- dataset_name - Any unique string for the name of the dataset
- contain_zero_values - Whether the dataset contains zero values(0/1)
- forecast_horizon - The forecast horizon of the dataset
- no_of_series - The number of series of the dataset
- optimizer - The type of the optimizer(cocob/adam/adagrad)
- seasonality_period - The seasonality period of the time series
- address_near_zero_instability - Whether the dataset contains zero values(0/1) - Whether to use a custom SMAPE function to address near zero instability(0/1). Default value is 0
- without_stl_decomposition - Whether not to use stl decomposition(0/1). Default is 0
- TFT
The codes in
src/models/TSMixer/tft_colab.ipynbshould be run in Colab instead, which is more likely to success in installing modules
There are three parts for evaluation.
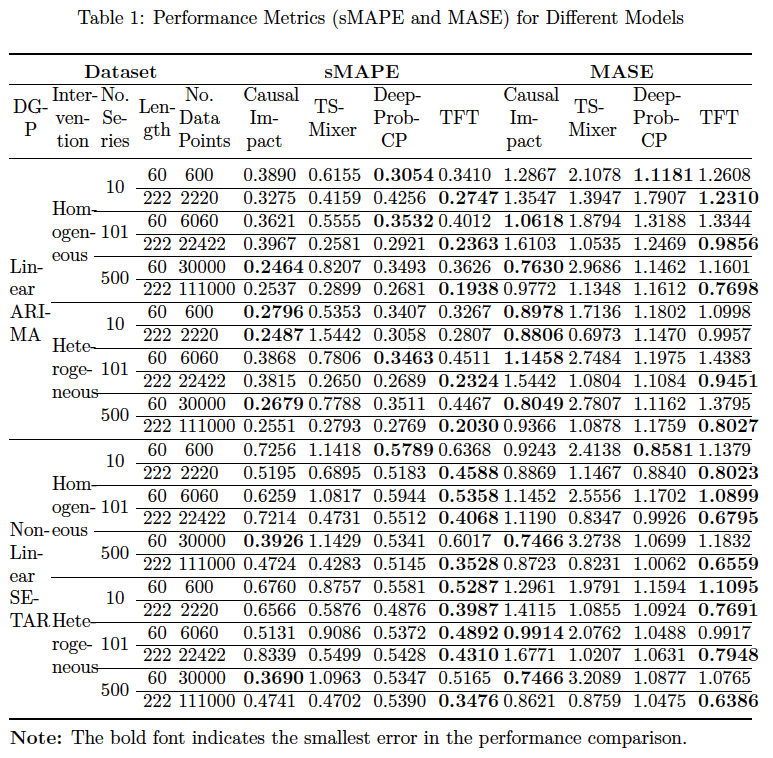
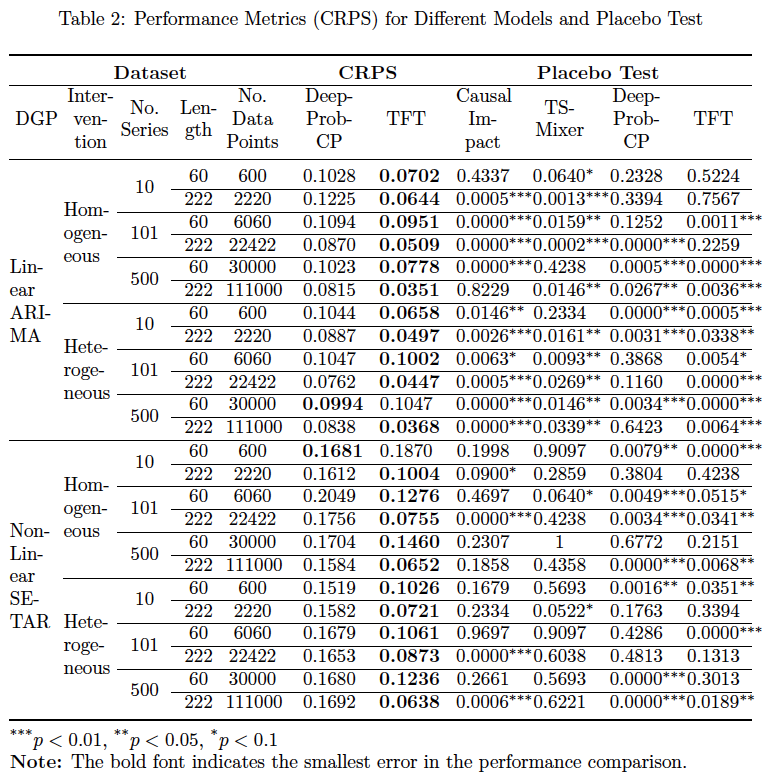
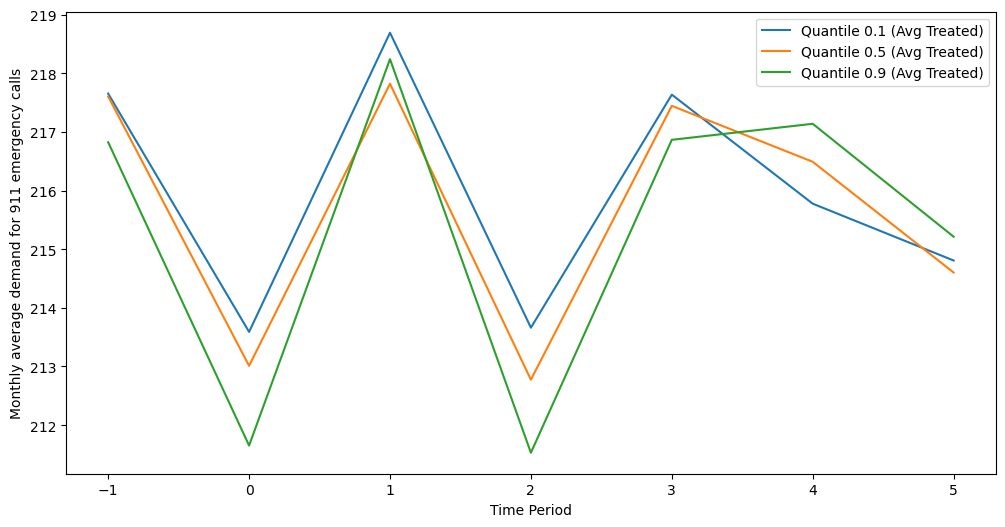
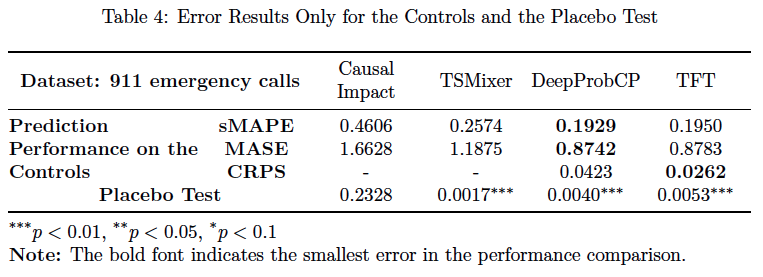
- The sMAPE, MASE and CRPS in the post-treatment period for the control and the treated (if the data is synthetic)
This is achieved during the modelling process, and the results are stored in results/.../accuracy
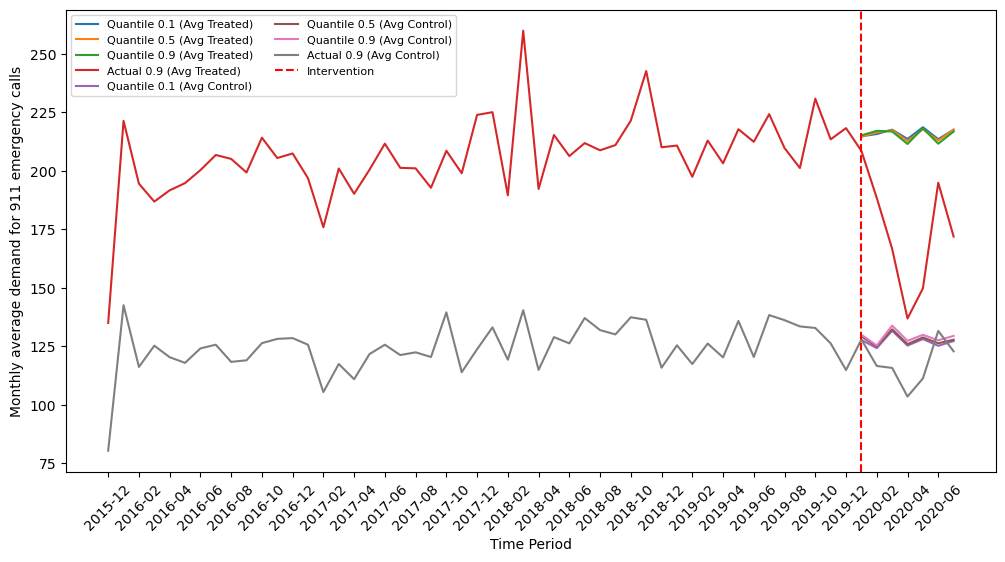
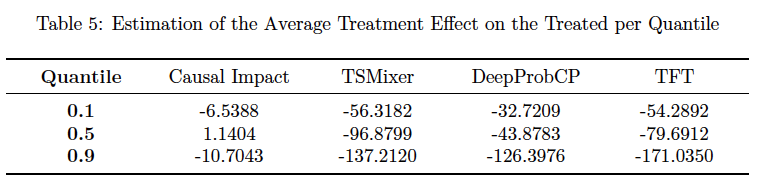
- The average treatment effect on the treated (ATT) in the post-treatment period for the treated and the sMAPE of ATT (if the data is synthetic)
This is executed in src/evaluation/results.ipynb, and the resultes are in results/.../tte
- Placebo Test
This is conducted and presented in
src/evaluation/results.ipynb
Pretrained models for DeepProbCP are in results/calls911/DeepProbCP/pkl and for TFT are in results/calls911/TFT/pkl
The main figures that are used in the thesis and the talk are in figures. For example, EDA_911.png is generated via src/prepare_source_data/calls911/EDA.ipynb. The others are mostly built in src/evaluation/results.ipynb. The numbers in tables are in results
├── README.md
├── requirements.txt -- required libraries
├── data -- stores txt and csv file
├── figures -- stores image files
├── results -- all results regarding predicted value, performance metric like sMAPE, MASE and CRPS, intermediate results and pretrained model and optimized hyperparameters
├── calls911
└── sim
└── src
├── prepare_source_data -- preprocesses data
├── calls911
├── [1]EDA.ipynb
├── [2]calls911_wrangling_code.R
├── [3]calls911_to_forecasting_code.R
├── [4]adjustOrigTestDtSet.R
├── [4]adjustOrigTrainDtSet.R
├── [5]create_tfrecords.py
├── [6]mean_stl_test_withoutEXvar.R
├── [6]mean_stl_train_validation_withoutEXvar.R
└── [7]benchmarks.py
└── sim
├── [1]simulation.html
├── [1]simulation.rmd
├── [2]preprocessing_layer.R
├── [3]create_tfrecords.py
└── ar_coefficients_generator.R
├── model -- modeling process
├── CausalImpact
└── causalimpact.ipynb
├── DeepProbCP
├── cocob_optimizer -- optimizer for DeepProbCP
├── configs -- configurations for DeepProbCP
├── error_calculator -- evaluation functions for DeepProbCP
├── quantile_utils -- quantile-related functions for DeepProbCP
├── rnn_architectures -- main file for model structure, the one with "_p" is for DeepProbCP
├── tfrecords_handler -- handles the .tfrecords data
├── [for R]invoke_final_evaluation.py -- original evaluation function from DeepCPNet
├── DeepProbCP.sh -- execution scripts for terminal
├── ensembling_forecasts.py -- the forecasts from the 10 seeds are ensembled by taking the median
├── generic_model_handler.py -- main function evoked by DeepProbCP.sh
├── hyperparameter_config_reader.py -- read the hyperparameter
├── peephole_lstm_cell.py -- peephole structure for lstm cell
└── persist_optimized_config_results.py -- configuration-related
├── TFT
└── tft_colab.ipynb
└── TSMixer
├── tsmixer_load -- data_loader for tsmixer to implement window moving strategy
└── tft_colab.ipynb
└── evaluation -- evaluate in terms of causal impact and placebo test
└── results.ipynb -- a giant but comprehensive notebook