Analysis of ACE2 Genetic Variability among Populations Highlights a Possible Link with COVID-19-Related Neurological Complications
This repository contains genetic data and R code we used in the Analysis of ACE2 Genetic Variability among Populations Highlights a Possible Link with COVID-19-Related Neurological Complications scientific article. The article was published in July 2020 by Genes, which is an international peer-reviewed scientific journal. The article is published in open access.
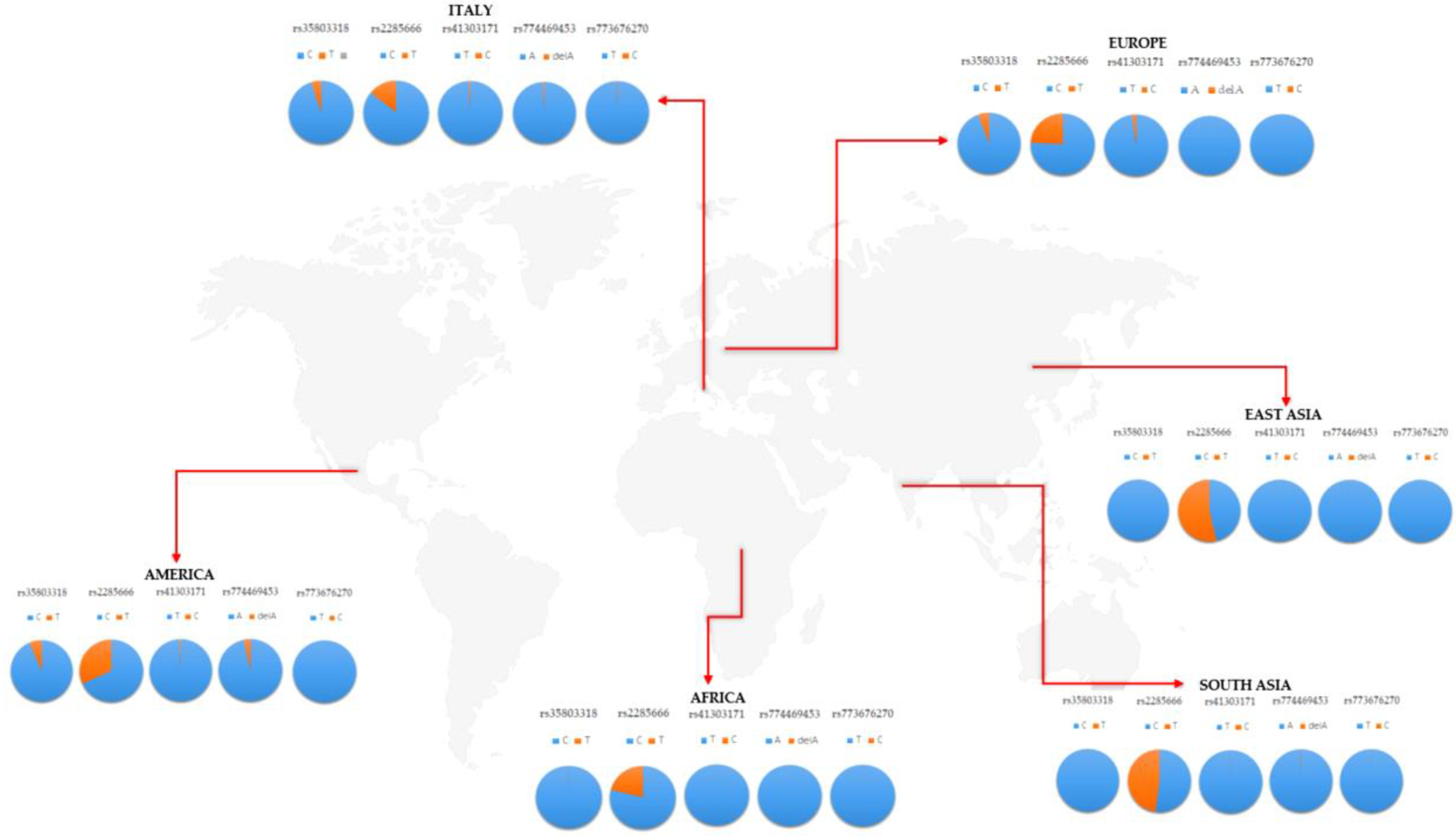
Angiotensin-converting enzyme 2 (ACE2) has been recognized as the entry receptor of the novel severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2). Structural and sequence variants in ACE2 gene may affect its expression in different tissues and determine a differential response to SARS-Cov-2 infection and the COVID-19-related phenotype. The present study investigated the genetic variability of ACE2 in terms of single nucleotide variants (SNVs), copy number variations (CNVs), and expression quantitative loci (eQTLs) in a cohort of 268 individuals representative of the general Italian population. The analysis identified five SNVs (rs35803318, rs41303171, rs774469453, rs773676270, and rs2285666) in the Italian cohort. Of them, rs35803318 and rs2285666 displayed a significant different frequency distribution in the Italian population with respect to worldwide population. The eQTLs analysis located in and targeting ACE2 revealed a high distribution of eQTL variants in different brain tissues, suggesting a possible link between ACE2 genetic variability and the neurological complications in patients with COVID-19. Further research is needed to clarify the possible relationship between ACE2 expression and the susceptibility to neurological complications in patients with COVID-19. In fact, patients at higher risk of neurological involvement may need different monitoring and treatment strategies in order to prevent severe, permanent brain injury.
This repository contains ACE2 genetic data of italian and worldwide population. Genetic data were retrieved from ENSEMBL API and 1000genomes project. The Italian cohort of samples was composed of 100 samples analyzed by a comparative genomic hybridization (aCGH) array for assessing the presence of structural genomic variations, and 168 samples utilized for identifying common and rare variants located in the coding or splice site regions of the genome. The genetic data referred to these samples were partially derived by whole exome sequencing (WES), available at the Genomic Medicine Laboratory of IRCCS Santa Lucia Foundation Hospital and partially extracted by the Ensembl database. Italian patients had an average age of 46 ± 15 years and a female/male ratio of 45:55. The research was approved by the Ethics Committee of Santa Lucia Foundation of Rome (CE/PROG.650, approved on 1 March 2018), and was performed according to the Declaration of Helsinki. The participants provided signed informed consent.
We implemented R to compare ACE2 genetic variability across worldwide population. A two-sided Fisher’s exact test and a p-value (p) were calculated in order to assess the different allelic distributions of detected SNVs in the Italian cohorts with respect to the other populations. The significance threshold was set at p < 0.05. In addition, multiple testing correction (false discovery rate) was performed by calculating the q-value (q) [22] and setting a significance cutoff at at q < 0.05. Considering that ACE2 maps to the X chromosome, statistical analysis (two-sided Fisher’s exact test) was also performed by stratifying the cohorts according to gender to evaluate sex-related effects. Moreover, the SNVs detected in the Italian population were subjected to bioinformatic predictive analysis to assess their potential impact on ACE2 protein function and splicing mechanisms.
You can find the article here
Strafella, C., Caputo, V., Termine, A., Barati, S., Gambardella, S., Borgiani, P., ... & Cascella, R. (2020). Analysis of ACE2 genetic variability among populations highlights a possible link with COVID-19-related neurological complications. Genes, 11(7), 741.