-
Notifications
You must be signed in to change notification settings - Fork 73
Tutorial 1 EEG to ERPset
We assume that Matlab is installed, that EEGLAB is installed and in the Matlab path, and that ERPLAB is correctly installed in the EEGLAB plugin subfolder. If not, see the installation instructions here.
| Requirements | Completed? |
|---|---|
| Load EEG Data | |
| Creating an EventList | |
| Assigning Trials to Bins | |
| Creating Bin-Based EEG Epochs | |
| Artifact Detection | |
| Creating Averaged ERPs |
Download the example EEG data files here. Unzip to a known folder, here we show C:\m\eegdata\erplab_tut. Note that there is a subfolder for each experiment subject, S1 to S6.

Within the S1 subfolder, you can see the S1_EEG.set file. This file contains the continuous EEG data for subject 1 in EEGLAB format, and also includes some meta data, like the event code timing information.
You should see something close to these screenshots. :
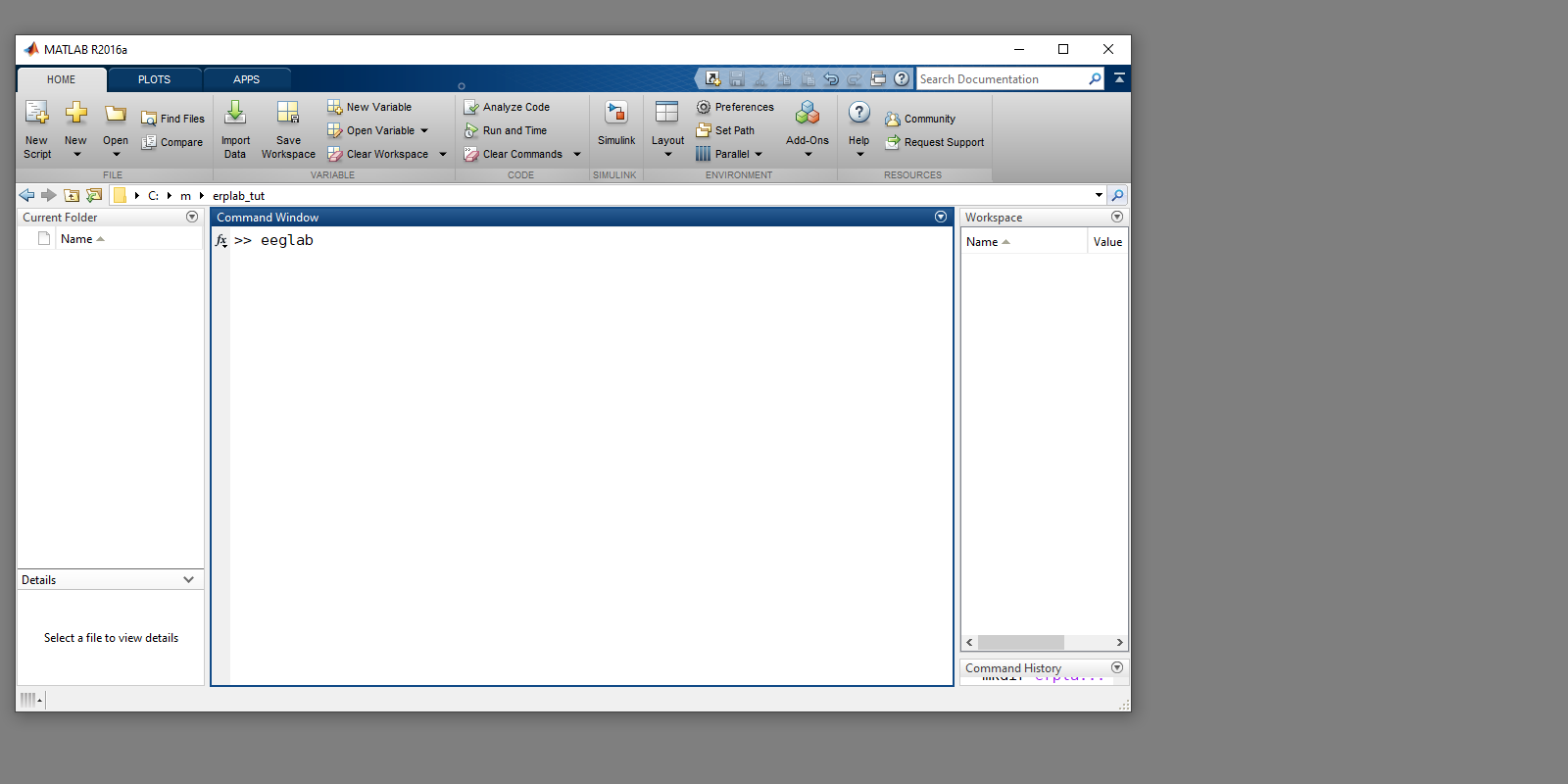
Matlab environment, entering eeglab in the Command Window
A new window appears for EEGLAB, with the ERPLAB menu:
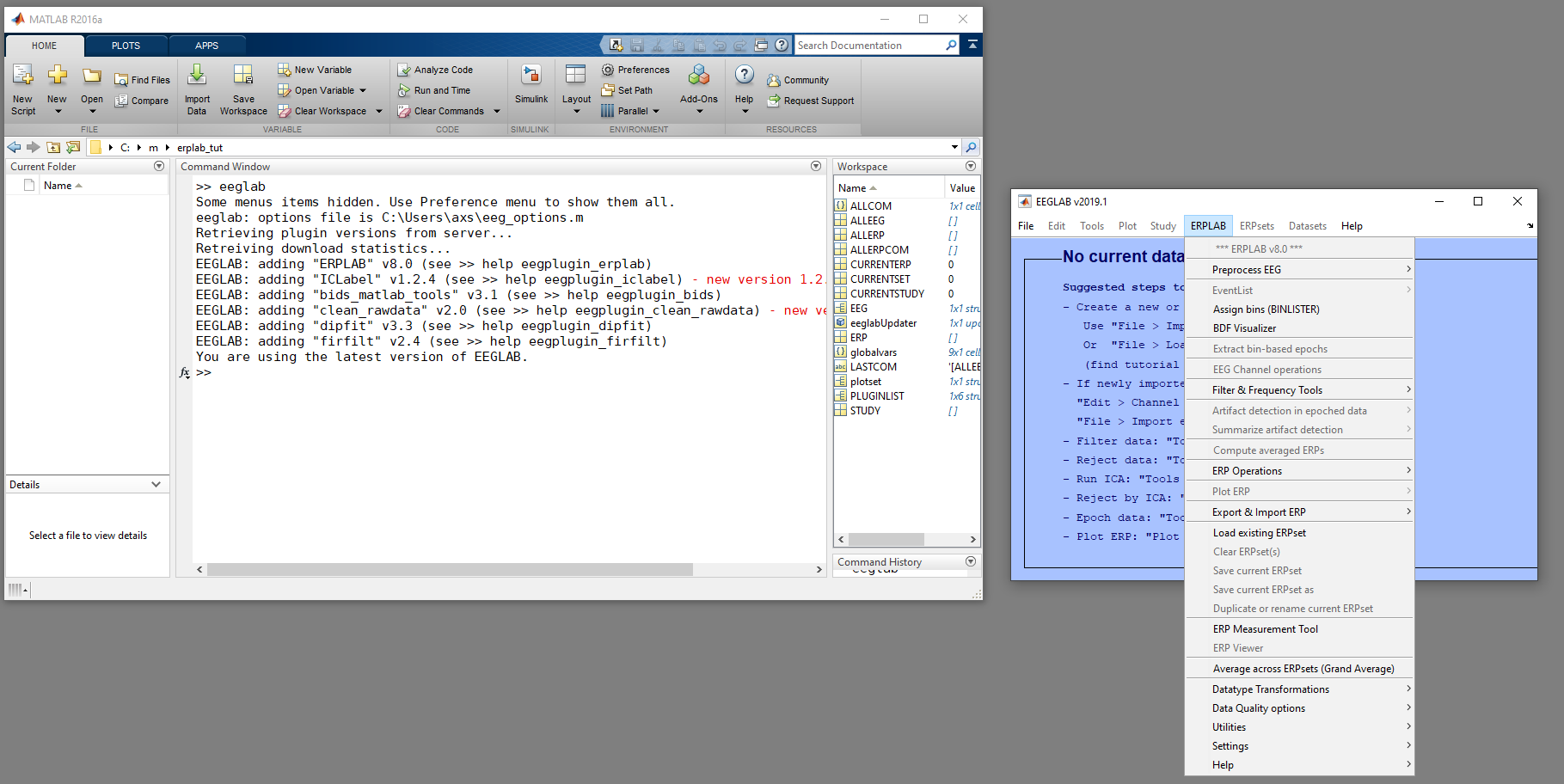
If you encounter an error here, check that ERPLAB is installed correctly, and the troubleshooting hints.
With EEGLAB started within Matlab, hit File > Load existing dataset.

Select the S1_EEG.set file, from the S1 subfolder in the path you unzipped example EEG data to.
The EEGLAB window should now update to show some information about this loaded EEG set:

Also, a list of currently loaded EEG sets is shown in the EEGLAB window as a dropdown menu, now containing Dataset 1: S1_EEG. There is also dropdown menu for ERPsets, and this remains empty, as we have only loaded continuous EEG data.
There are several aspects to the EEG datasets that are good to keep in mind. As these can be observed and verified here, it is good practice to do so at this stage:
- What data do I have? How many EEG channels? What is the duration of the recording?
This can be found within the blue EEGLAB GUI (as shown in screenshots above), where the channels per frame tells you the number of channels. The number of epochs is 1, the epoch start time is 0, and the epoch end time is 2139 s, indicating that the loaded and currently selected EEG set is continuous data, and runs from t=0 s to t=2139 s, giving (2139/60) around a 35 minute recording. This is EEG data recorded at a sample rate of 500 Hz, so there are (2139 * 500) around 1,069,500 datapoints for each electrode.
This information can also be accessed programatically within Matlab, with commands like:
EEG.nbchan size(EEG.data) EEG.srate
- What events occur within this recording? Are there event codes (aka event markers, triggers) indicating the relevant times of stimuli presentation and response?
From the EEGLAB window, we can plot the EEG recording data, through Plot menu -> Channel data (scroll).

Channel data is presented with one channel atop another, with channel F3 on top here. If you advance the time from 0 s to 10 s, you can observe two large deflections of more that 50 µV that occur at around 10.5 s and 12 s. These are likely blinks.

Additionally, there are straight vertical dashed lines, of different colors. There are none of these in the first 12 seconds of this recording, and 4 in the trace shown above, at around 13.1, 13.6, 14.3, and 14.8 seconds. These are the event codes, indicating an event of type 22, 9, 12, 9 occurred at the respective times.
These event codes have been generated by the experimental stimuli script at the time of recording the EEG, and included with the EEG data. The 22 indicates the presentation of a certain type of frequently shown stimulus, the 12 indicates a certain type of rarely shown stimulus, and 9 indicates the time at which the subject made a buttonpress response. Here, we wish to observe that this data exists, and agrees with our expectations of this experiment.
This information can also be accessed programatically within Matlab, with commands like:
pop_squeezevents(EEG)

- What ERPs are desired? What timeframe is desired to show these ERPs?
Your research question will determine what the ERP and timeframes that are central for your experiment. Hopefully, your experimental design captures this ERP component well. There exists guides to help, like the Oxford Handbook of ERPs for example.
Now that we have loaded and examined some data, the next step is to attach an EventList:
| Requirements | Completed? |
|---|---|
| Load EEG Data | ✓ |
| Creating an EventList | |
| Assigning Trials to Bins | |
| Creating Bin-Based EEG Epochs | |
| Artifact Detection | |
| Creating Averaged ERPs |
At the moment, this dataset is continuous EEG data, with event markers indicating the times of trial-relevant stimulus presentation.
In order to turn this data into an Averaged ERP dataset, we need to keep track of these events in an ERPLAB format EventList, and then determine what events should go in what bin of the ERP.
Here, we wish to group the Frequent stimuli type together in one bin, and the Rare stimuli type in another, to show the difference in their averages in a P300 ERP. For more information about the experiment, see here.
The EventList is a refactoring of the trial information in a more ERPLAB-interpretable way, so we can address these events later on.
The author of this experiment has used the following stimuli event codes to mean:
Event Code Category Probability Correct Response
11 Letter Frequent Left Hand
21 Digit Rare Right Hand
112 Letter Rare Left Hand
122 Digit Frequent Right Hand
12 Letter Rare Right Hand
22 Digit Frequent Left Hand
111 Letter Frequent Right Hand
121 Digit Rare Left Hand
To obtain counts of the number of times each event is present in the active EEGset, you can enter pop_squeezevents(EEG).
To create an EventList for the EEG data we loaded above, let's hit
ERPLAB menu > EventList > Create EEG EVENTLIST

That will pop up this new window. Optionally, we can write the EL to a text file. The information will still be 'attached' to this EEGset, but that text file can be useful for examining outside Matlab.
Note that the EventList does not like strings. Event names are best to be numbers and not text. If your event names are text, we suggest converting them to a numeric code here. The events here are all numbers, but stored as stings for now. Using the 'Create numeric equivilent' option in the 'Create EventList' GUI, the text is dropped from a string event code, leaving the number. This option will trim any text, and convert to numeric type, and so will work for these event codes. If you need to convert different types of string events to numeric, you can use the 'Convert string code' editable option in the middle of this window, but this is not needed for this example dataset.
The boundary code is an important event code, indicating the presence of a discontinuity, like a removed section of data, or a break in the recording. We recommend keeping that in the data with the numeric indicator of event -99.

After hitting 'CREATE', a new EEG set is created -- same as the previous one, but now with a EventList attached.
Let's save this new EEG set in the S1 folder.
Optionally, to inspect this new EL, we can probe
EEG.EVENTLIST.eventinfo
or explore there in the Matlab Workspace.

If you chose to write the EventList to a text file, you can also view it in the Matlab editor with something like:
edit S1/EventList_1.txt
This shows our events (all 2557 of them), and the time that they occurred.
Note that at the moment, the code field is filled, and that the binlabel field is empty.
| Requirements | Completed? |
|---|---|
| Load EEG Data | ✓ |
| Creating an EventList | ✓ |
| Assigning Trials to Bins | |
| Creating Bin-Based EEG Epochs | |
| Artifact Detection | |
| Creating Averaged ERPs |
With the events coded in the EventList, we can now specify what we want in ERP bins.
We have event codes for different 'frequent' or 'rare' stimuli presentation times. For this P300 experiment, we wish to group all the 'frequent' stimuli in to one bin, and the rare in to another, so they can then be averaged together and compared in in ERP set.
From the design of this experiment, we know that all of {11,122,22,111} event codes indicate Frequent stimuli, and that all of {21,112,12,121} are Rare stimuli. Additionally, an event code of {9} indicates a correct response.
The syntax of BINLISTER allows specification of bins for complex experiments. Here, we will go for a simple example, but I hope I can show you how it might be powerful.
The idea is to set instruction for each bin in a specific format, called the Bin-Descriptor Files:
bin 1
Frequent followed by correct response
.{11;122;22;111}{9}
bin 2
Rare followed by correct response
.{21;112;12;121}{9}
These Bin-Descriptor Files are saved as raw text files, and used by ERPLAB's BinLister function to specify ERP bins. You can see an example of these files in the Test_Data/binlister_demo_1.txt file. Open this file with the Matlab text editor by right-clicking it in the Matlab 'Current Folder' section, or through edit binlister_demo_1.txt if you are in the Test_Data directory. Using MS Word to edit these files can add incorrect formatting, so we recommend using the Matlab editor (or, perhaps, another raw text editor like Sublime Text.
Note that there are exactly 3 lines for each bin, followed by an empty line, like:
bin <number_n>
Text name of bin
.{1;2}{3}
Let's unpack that.
The bin numbers start at 1, and go up by 1.
The text name of the bin is specified by you. Make it unique and descriptive.
The event specification is in the 3rd line. Each pair of curly brackets {} defines an event set, which contains one or more event codes. One of the event sets is preceded by a period, which indicates that this event set is the time-locking event for that bin (i.e., the event occurs at time zero).
You can include multiple event codes within an event set by separating them with semicolons. For example, {1;2} means that an event code of 1 or an event code of 2 are valid for this bin.
When there are multiple event sets (multiple sets of events in curly brackets), this allows you to specify that a specific sequence of events must be found. For example, .{1;2}{3} means that an event code 1 or 2 must be IMMEDIATELY followed and event code 3. For example, this description would be matched by event code 1 immediately followed by event code 3 or event code 2 immediately followed by event code 3, but it would not be matched by a sequence of 1 followed by 5 followed by 3.
With both an EventList and Bin-Descriptor file, we can run BINLISTER to apply that Bin-Descriptor to the loaded EEG dataset.
ERPLAB menu -> Assign bins (BINLISTER)

In the top section, hit 'Browse' and select the binlister_demo_1.txt file. Optionally, you can hit 'Take a look' to verify that this is the bin-descriptor you want.
On hitting 'RUN' on this window, BinLister will examine all the events, and, when there is a match to any of the requested bins, add this information to the EventList. This will create a new EEG dataset, now with the suggested name appended with '_bins', so 'S1_EEG_elist_bins' by default for our first subject here.
If we examine the EEG.EVENTLIST.eventinfo structure in this new EEG data in the Matlab Workspace variable explorer, we can see the updated EventList, now with bin info. For instance, the binlabel field for the first item now shows that it will be grouped to Bin 1, as it has a 22 event code that is immediately followed by a 9. The binlabel for the third item shows it has been has a Bin 2 label, as it was a 12 event code followed by a 9.
| Requirements | Completed? |
|---|---|
| Load EEG Data | ✓ |
| Creating an EventList | ✓ |
| Assigning Trials to Bins | ✓ |
| Creating Bin-Based EEG Epochs | |
| Artifact Detection | |
| Creating Averaged ERPs |
Now that the bin information is present, we can convert this continuous EEG dataset into a series of short stretches of data oriented around key events, making an epoched EEG set. For all periods of the EEG data that match one of the above-defined bins, we will take a period of time around the defined event (typically around 200 ms before and 800 ms after), and that will be an epoch. Epochs will be anchored around specific event markers, with some time before and after. Rather than the previous continuous EEG data stored in the format of
(number of electrodes) * (number of time-points)
epoched EEG data will now be stored in the format of
(number of electrodes) * (number of time-points in epoch) * (number of epochs/trials)
Running the command of size(EEG.data) on our existing EEG set reveals a 16 electrode by 1 million datapoint 2D data array.
To extract bin-based epochs, hit:
ERPLAB menu -> Extract bin-based epochs

We specify the time range in the top section -- here, -200 up to 800 ms. This means each epoch will be 1000 ms long. Note that this is in milliseconds, relative to the event specified with a leading period in the bin-descriptor file, like .{11}.
Hitting 'Run' here will create a new EEG dataset, with the now-epoched data. To indicate that this is bin-epoched data, the suggested name of the EEG set will be appended with '_be', giving S1_EEG_elist_bins_be if you have followed all steps so far.
You can confirm that this data is now epoched by seeing that the blue EEGLAB window now lists 'Epochs: 1251'. This tells us that from the around 2500 events present in this EEG dataset, there are 1251 matches for the bins that we specified with the BDF above.

Additional confirmation can be seen by entering the command of size(EEG.data) again, reveals a 16 electrode by 500 datapoints by 1251 epoch 3D data array. Note that 1 second of data at 500 Hz gives 500 datapoints per epoch.
| Requirements | Completed? |
|---|---|
| Load EEG Data | ✓ |
| Creating an EventList | ✓ |
| Assigning Trials to Bins | ✓ |
| Creating Bin-Based EEG Epochs | ✓ |
| Artifact Detection | |
| Creating Averaged ERPs |
At this point, it would be possible to average these epochs into their respective bins, forming an ERP set. Before doing so, we can improve the data quality of our ERPs by marking epochs that have obvious artifacts.
For example, if we look at the first 5 epochs of the now-epoched EEG trace (through Plot menu -> Channel Data scroll), we can see that a large deflection exists at around 550 ms in to epoch 5. This is probably due to the subject blinking. We can see that there are also such artifacts in epoch 8 and 11, and in more epochs too. If we were to average this data without marking and excluding these epochs, these large non-neural deflections would add to the noise of our ERP results.

ERPLAB has several tools for detecting and marking different types of artifacts. Note that these tools will mark epochs as identified as being an artifact. This does not remove data from the EEG set, but instead allows these to then easily be excluded from the epochs that go into an ERP bin. This is contrasted with some EEGLAB artifact rejection functions, which will delete and remove sections from the EEG set.
If you do perform EEGLAB artifact rejection, be sure to regenerate a ERPLAB EventList afterwards.
Many types of artifacts can be detected with ERPLAB's moving window peak-to-peak tool.
Hit ERPLAB menu -> Artifact detection in epoched data -> Moving window peak-to-peak threshold
This will open a new window, where we can specify parameters for this artifact detection method. This function will has a small window of analysis that steps through the time if the epochs, and records the difference between the point of lowest and highest amplitude. If this is ever above a stated threshold, the whole epoch is marked as an artifact. For more details, hit ? in the window, or see the ERPLAB manual page here.

By default, the 'test period' will be the whole epoch, stated in milliseconds. We go with the default moving window size of 200 ms, with 100 ms step size. The 'Voltage Threshold [µV]' is a key parameter, that may change across your subjects. For more sensitivity, lower this threshold, and more epochs will be found to be above this. For capturing the large blinks in S1 here, a value of 100 µV might be sensible.
Note that there are also 'flags' which can be used to disambiguate what artifact detection tools marked which epochs. The F1 flag is active for all artifacts, and, optionally, you can select others too. Here, Let's press the flag 2 button, so that both F1 and F2 are marked.
Hitting 'Accept' here will run this artifact detection tool with these parameters, and create a new epoched EEG set with those matching epochs marked as artifacts. By default, '_ar'will be appended to the EEG set name, giving 'S1_EEG_elist_bins_be_ar' for our new EEG set.
ERPLAB will also output a summary of the artifact detection operation:

Note that 288 epochs are now tagged as artifacts from Bin 1, and 72 from Bin 2. This is around 29% of trials from each bin now tagged as artifacts (which is a large but not crazy fraction), leaving 721 trials with now artifact tag in Bin 1, and 170 in Bin 2. You can examine this artifact summary again through pop_summary_AR_eeg_detection(EEG); or ERPLAB menu -> Summarize artifact detection .
Examining the EEG trace after this artifact detection process (through EEGLAB Plot menu -> Channel data (scroll)) reveals that many epochs with large artifacts are now highlighted in yellow. That includes the epoch 5, 8, and 11, which we previously noted as having likely blink artifacts.

Additional artifact detection routines can be run to find more artifacts, and have more accurate detection. See the ERPLAB manual page here.
| Requirements | Completed? |
|---|---|
| Load EEG Data | ✓ |
| Creating an EventList | ✓ |
| Assigning Trials to Bins | ✓ |
| Creating Bin-Based EEG Epochs | ✓ |
| Artifact Detection | ✓ |
| Creating Averaged ERPs |
The final step here is to actually run the Averager, taking this processed epoched EEG set, and outputting and Averaged ERP dataset.
Hit:
ERPLAB menu -> Compute averaged ERPs

At the top, we see the selected EEG dataset is number 5, which corresponds to the active EEG dataset here - S1_EEG_elist_bins_ar.
In the central section here, we specify that we wish to exclude epochs marked as artifacts. There are also options for selecting different sets of epochs, but we want all non-artifact epochs here. There is also an option for excluding any epoch that has boundaries or invalid events. We recommend having this tickbox enabled, so as to ignore epochs with boundary edge effects.
By default, we also have automatic data quality assessment enabled, with default options. This will include information about baseline noise, SEM, and Standardized Measurement Error (aSME) in the ERPset (see more details for these options here).
Hitting 'Run' here will create a ERP dataset.
This will pop up the 'Save ERPset GUI' for the first time. Let's save this as 'S1_ERP', and also save this new ERPset to a file, within the S1 subject folder:

With this new ERPset created, we now have an active ERPset that is selected. Alongside the EEGLAB 'Datasets' menu, which shows the 5 EEGsets from S1, we also have an 'ERPsets' menu, which shows the loaded ERPsets:
ERPset 1: S1_ERP

Averaging each bin in the above bin-epoched EEG data give us ERP data in the format:
(number of electrodes) * (number of time-points in epoch) * (number of bins)
With the data prepared in this ERP set, we can access and plot the data from each condition:

| Requirements | Completed? |
|---|---|
| Load EEG Data | ✓ |
| Creating an EventList | ✓ |
| Assigning Trials to Bins | ✓ |
| Creating Bin-Based EEG Epochs | ✓ |
| Artifact Detection | ✓ |
| Creating Averaged ERPs | ✓ |
We have a loaded ERPset! We can now run the many ERPLAB functions that act on this ERPset, including plotting and measuring tools.
We demonstrate use of some of these tools in the next tutorial.
|
Tutorial Overview |
Tutorial list |
Tutorial 2: ERP Plotting and Measuring |