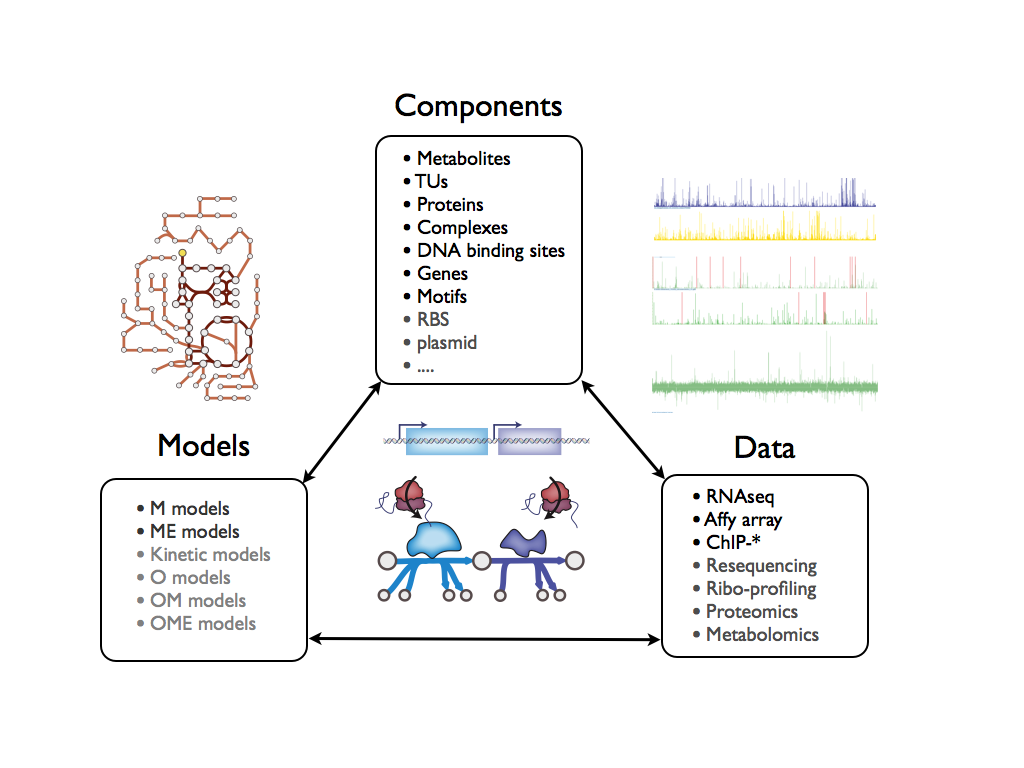
COBRAdb is a database for COBRA models, genomes, and biological datasets.
- map highthroughput experimental data to cellular components
- map cellular components to computational models
- map computational models to highthroughput experimental data via cellular components
The framework is written entirely in python with great help from SQLalchemy. You can browse the schemas below to get a sense of the underlying database design or jump straight to the examples page. Enjoy!
Most biological analyses and analysis tools are focused on working with a relatively limited subset of data concerned with a specific study or question of interest. However, as the sequencing and synthesis of DNA surges ahead it becomes clear that we will soon generate 10 replicates instead of 3, cross reference 10 historic datasets instead of 1, and most importantly begin to get a level of quantitation that will enable true biological engineering.
COBRAdb is a small step towards a future that amasses, manages, and processes biological data at a systems scale.