#Neuron Finding
Team name - minions
Team members: Chirag Jain, Sharmin Pathan
Approach: NMF and median filtering to find neurons in calcium imaging data
Python 2.7
Thunder
Apache Spark
Matplotlib
Median Filtering
NMF
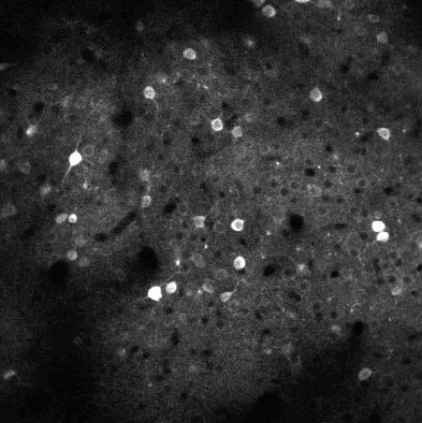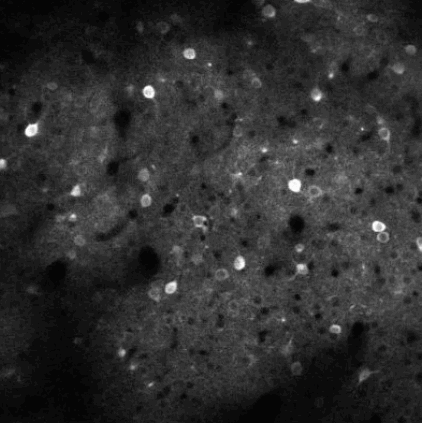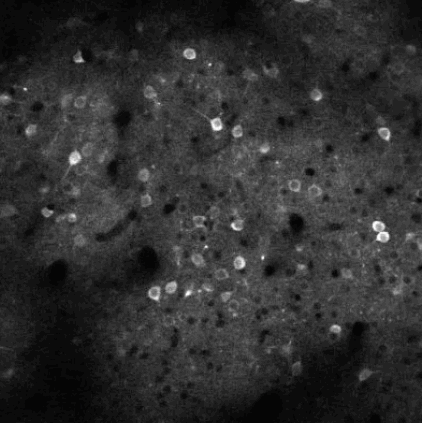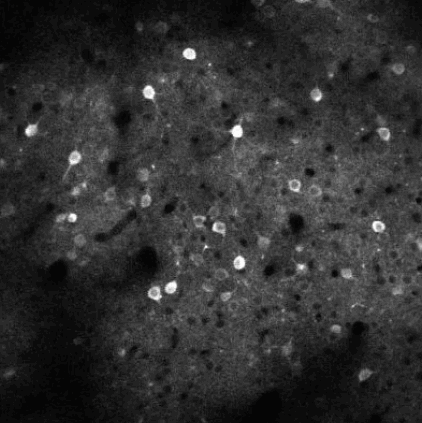
Calcium Imaging is a dominant technique in modern neuroscience for measuring activity of large population of neurons. Several challenges remain in how to best process and extract signals from these data.
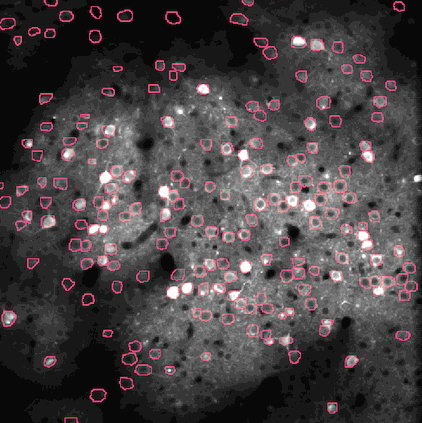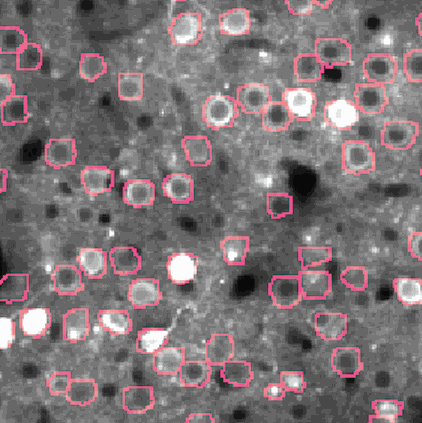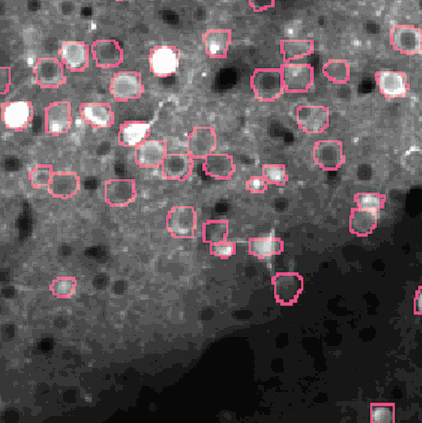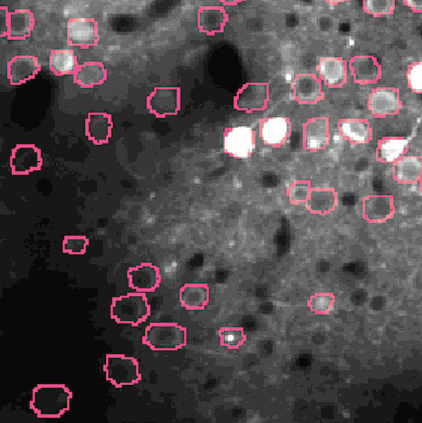
The challenge is to take the time varying images (left) and extract regions of interest (right) that correspond to individual neurons.
The datasets were taken from http://neurofinder.codeneuro.org/ which includes 19 training sets and 9 test sets.
- A structured matrix factorization approach to analyzing calcium imaging recordings of large neuronal ensembles.
- Demix spatially overlapping regions
- Denoise and deconvolve spiking activity of each neuron
- Median filtering of the images
-
Load the datasets
-
NMF (Non-negative matrix factorization) to express the spatiotemporal fluorescence activity as a product of two matrices: a spatial matrix that encodes location of each neuron, and a temporal matrix that characterizes the calcium concentration of each neuron over time.
-
The NMF algorithm is contructed using the following attributes
- k=5: number of components to estimate per block
- max_iter=50: maximum number of algorithm iterations
- percentile=99: value for thresholding (higher means more thresholding)
- overlap=0.1: value for determining whether to merge (higher means fewer merges)
-
The algorithm is then fit to the data. The algorithm is applied to chunks of data and not the entire data with a required padding. We kept the chunk size to be between 30-60 and padding of 15x15 or 32x32.
-
Merge overlapping regions that are similar to one another more than the specified overlap.
-
A list of the identified regions is created.
-
The identified regions list is then saved into a json in the required format for submission.
- Ensure the following are installed
- Thunder
- Apache Spark
- Matplotlib
- The program takes four command line arguments, namely
- name of the dataset
- no. of components to estimate per block
- chunk size
- padding
- For example: python neuro.py 00.00.test 5 50 25
#####We applied different chunk sizes and padding for different datasets since their sizes varied in terms of the no. of images.
| Dataset | k | Chunk size | Padding | Regions |
|---|---|---|---|---|
| 00.00 | 5 | 30x30 | 15x15 | 356 |
| 00.01 | 32 | 32x32 | 32x32 | 503 |
| 01.00 | 32 | 32x32 | 32x32 | 326 |
| 01.01 | 32 | 32x32 | 32x32 | 264 |
| 02.00 | 5 | 100x100 | 0x0 | 108 |
| 02.01 | 5 | 100x100 | 0x0 | 099 |
| 03.00 | 5 | 60x60 | 15x15 | 439 |
| 04.00 | 5 | 60x60 | 0x0 | 201 |
| 04.01 | 5 | 30x30 | 15x15 | 331 |
- Tried coverting the images to grayscale. But, on studying the NMF algorithm and the datasets, we got to know that the images were already into grayscale format with channel=1
- Performed median filtering but that resulted into less no. of regions. So, we removed it from our final code.
- Image registration to align multiple scenes into a single integrated image. We studied over this topic but could not actually implement it due to lack of time.
- Made a submission on codeneuro
- It was difficult to come up with possible image preprocessing techniques that could be applied.
- Understanding Calcium Imaging and related sciences.