-
Notifications
You must be signed in to change notification settings - Fork 7
/
brainglobe-napari-io.json
173 lines (173 loc) · 9.77 KB
/
brainglobe-napari-io.json
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
{
"name": "brainglobe-napari-io",
"display_name": "brainglobe-napari-io",
"visibility": "public",
"icon": "",
"categories": [],
"schema_version": "0.1.0",
"on_activate": null,
"on_deactivate": null,
"contributions": {
"commands": [
{
"id": "brainglobe-napari-io.brainreg_read_dir",
"title": "Brainreg Read Directory",
"python_name": "brainglobe_napari_io.brainreg.reader_dir:brainreg_read_dir",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "brainglobe-napari-io.brainreg_read_dir_atlas_space",
"title": "Brainreg Read Directory Atlas Space",
"python_name": "brainglobe_napari_io.brainreg.reader_dir_atlas_space:brainreg_read_dir_atlas_space",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "brainglobe-napari-io.brainmapper_read_dir",
"title": "Brainmapper Read Directory",
"python_name": "brainglobe_napari_io.brainmapper.brainmapper_reader_dir:brainmapper_read_dir",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "brainglobe-napari-io.cellfinder_read_xml",
"title": "Cellfinder Read XML",
"python_name": "brainglobe_napari_io.cellfinder.reader_xml:cellfinder_read_xml",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "brainglobe-napari-io.cellfinder_write_multiple_xml",
"title": "Write Points",
"python_name": "brainglobe_napari_io.cellfinder.writer_xml:write_multiple_points_to_xml",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
}
],
"readers": [
{
"command": "brainglobe-napari-io.brainreg_read_dir",
"filename_patterns": [
"*.tiff"
],
"accepts_directories": true
},
{
"command": "brainglobe-napari-io.brainreg_read_dir_atlas_space",
"filename_patterns": [
"*.tiff"
],
"accepts_directories": true
},
{
"command": "brainglobe-napari-io.brainmapper_read_dir",
"filename_patterns": [
"*.tif"
],
"accepts_directories": true
},
{
"command": "brainglobe-napari-io.cellfinder_read_xml",
"filename_patterns": [
"*.xml"
],
"accepts_directories": false
}
],
"writers": [
{
"command": "brainglobe-napari-io.cellfinder_write_multiple_xml",
"layer_types": [
"points+"
],
"filename_extensions": [
".xml"
],
"display_name": "multiple_points"
}
],
"widgets": null,
"sample_data": null,
"themes": null,
"menus": {},
"submenus": null,
"keybindings": null,
"configuration": []
},
"package_metadata": {
"metadata_version": "2.1",
"name": "brainglobe-napari-io",
"version": "0.3.4",
"dynamic": null,
"platform": null,
"supported_platform": null,
"summary": "Read and write files from the BrainGlobe computational neuroanatomy suite into napari",
"description": "# napari-brainglobe-io\n\n[](https://github.com/brainglobe/brainglobe-napari-io/blob/main/LICENSE)\n[](https://pypi.org/project/brainglobe-napari-io)\n[](https://python.org)\n[](https://github.com/brainglobe/brainglobe-napari-io/actions)\n[](https://codecov.io/gh/brainglobe/brainglobe-napari-io)\n\nVisualise cellfinder and brainreg results with napari\n\n\n----------------------------------\n\n\n## Installation\nThis package is likely already installed\n(e.g. with cellfinder, brainreg or another napari plugin), but if you want to\ninstall it again, either use the napari plugin install GUI or you can\ninstall `brainglobe-napari-io` via [pip]:\n\n pip install brainglobe-napari-io\n\n## Usage\n* Open napari (however you normally do it, but typically just type `napari` into your terminal, or click on your desktop icon)\n\n### brainreg\n#### Sample space\nDrag your [brainreg](https://github.com/brainglobe/brainreg) output directory (the one with the log file) onto the napari window.\n\nVarious images should then open, including:\n* `Registered image` - the image used for registration, downsampled to atlas resolution\n* `atlas_name` - e.g. `allen_mouse_25um` the atlas labels, warped to your sample brain\n* `Boundaries` - the boundaries of the atlas regions\n\nIf you downsampled additional channels, these will also be loaded.\n\nMost of these images will not be visible by default. Click the little eye icon to toggle visibility.\n\n_N.B. If you use a high resolution atlas (such as `allen_mouse_10um`), then the files can take a little while to load._\n\n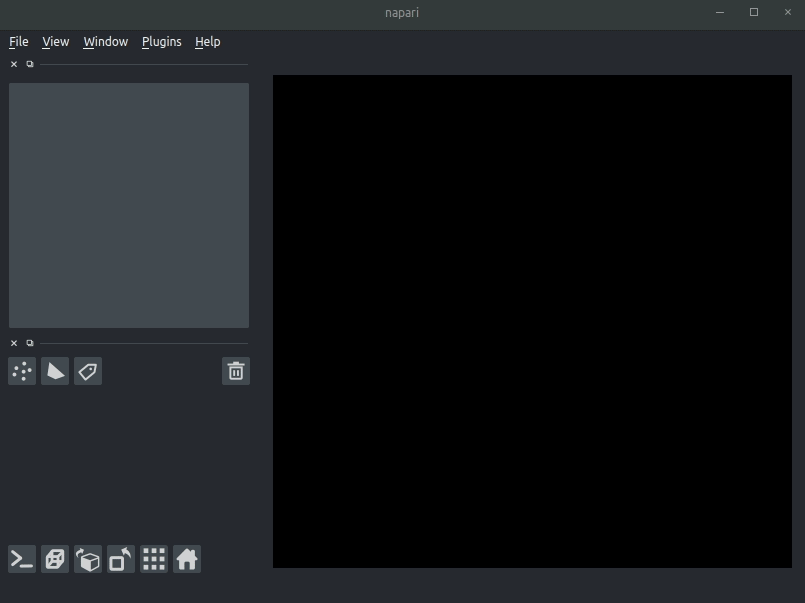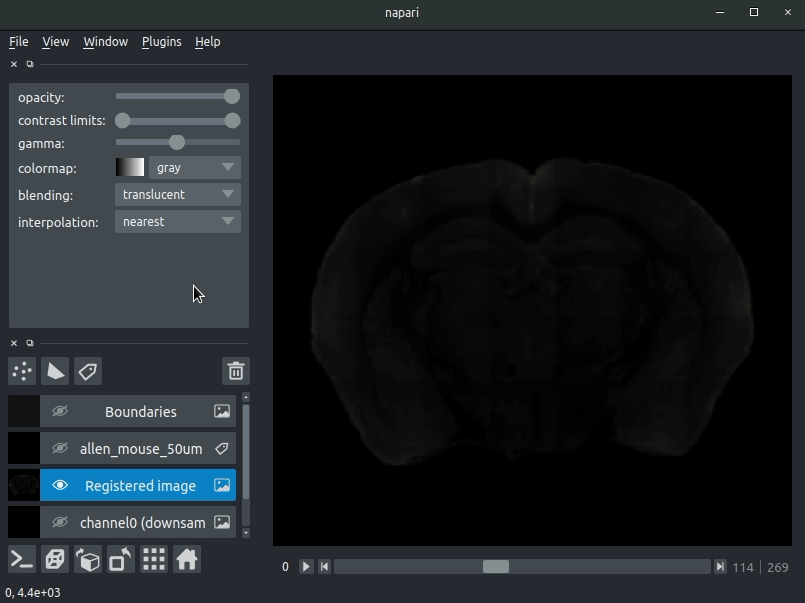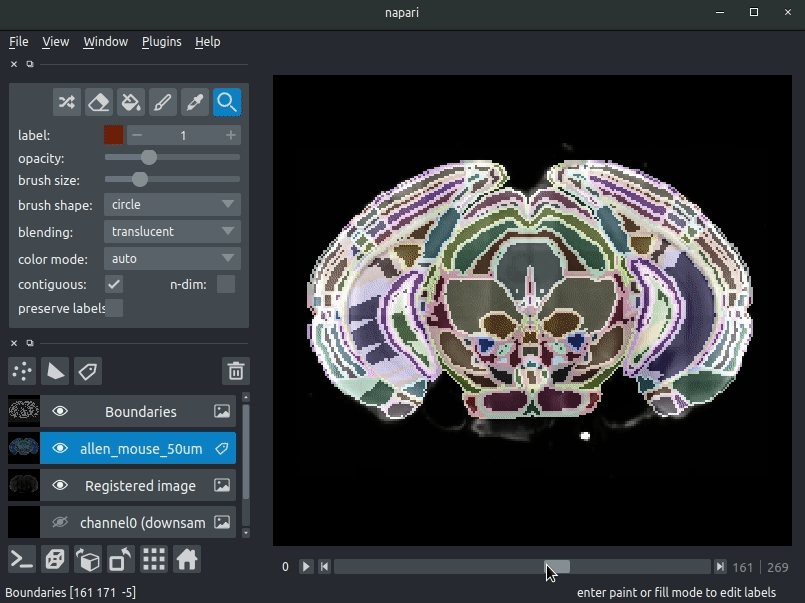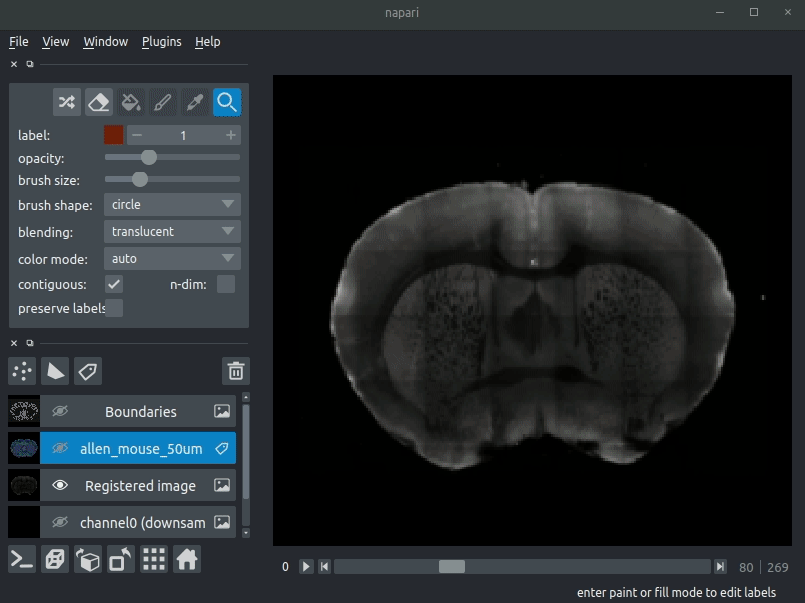\n\n\n#### Atlas space\n`napari-brainreg` also comes with an additional plugin, for visualising your data\nin atlas space.\n\nThis is typically only used in other software, but you can enable it yourself:\n* Open napari\n* Navigate to `Plugins` -> `Plugin Call Order`\n* In the `Plugin Sorter` window, select `napari_get_reader` from the `select hook...` dropdown box\n* Drag `brainreg_read_dir_atlas_space` (the atlas space viewer plugin) above `brainreg_read_dir` (the normal plugin) to ensure that the atlas space plugin is used preferentially.\n\n\n### cellfinder\n#### Load cellfinder XML file\n* Load your raw data (drag and drop the data directories into napari, one at a time)\n* Drag and drop your cellfinder XML file (e.g. `cell_classification.xml`) into napari.\n\n#### Load cellfinder directory\n* Load your raw data (drag and drop the data directories into napari, one at a time)\n* Drag and drop your cellfinder output directory into napari.\n\nThe plugin will then load your detected cells (in yellow) and the rejected cell\ncandidates (in blue). If you carried out registration, then these results will be\noverlaid (similarly to the loading brainreg data, but transformed to the\ncoordinate space of your raw data).\n\n\n**Loading raw data**\n\n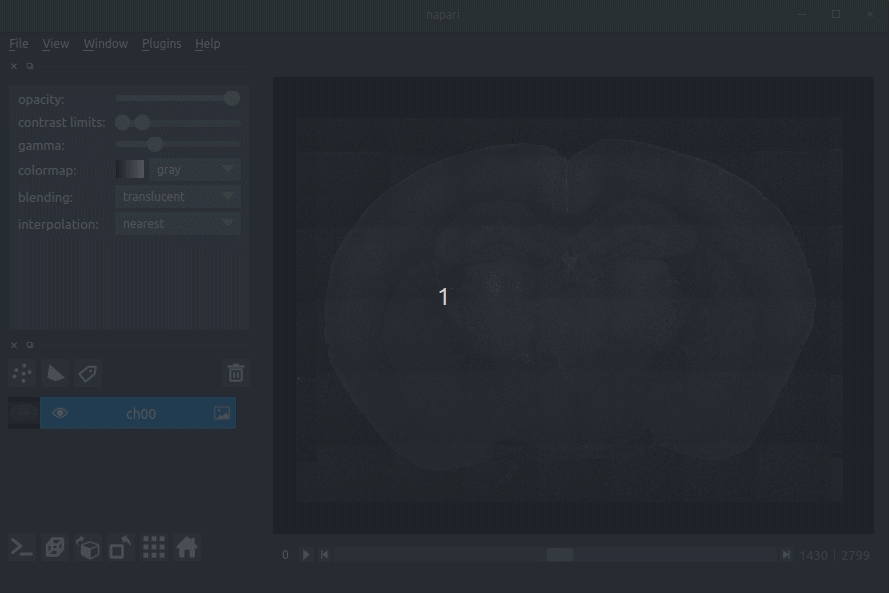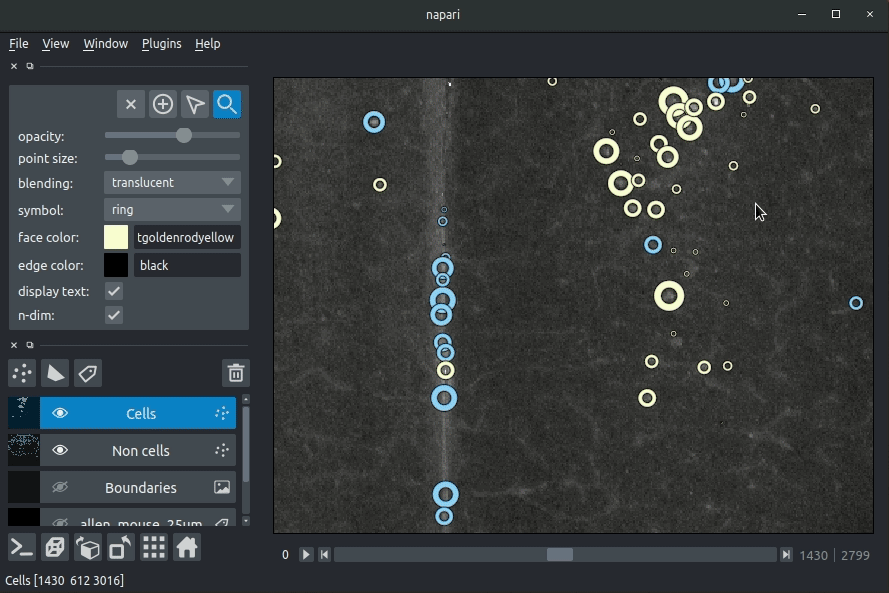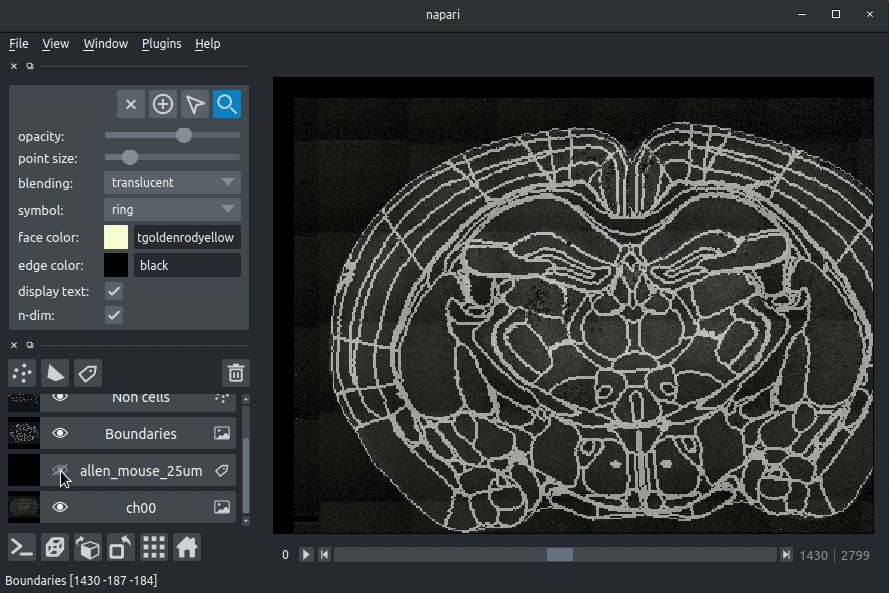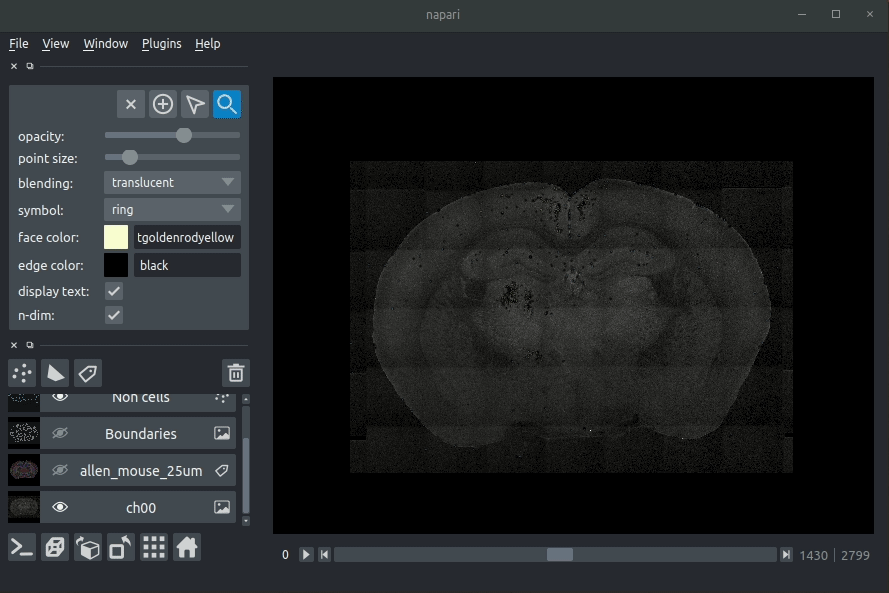\n**Loading cellfinder results**\n\n\n\n## Contributing\nPlease see the [developers guide](https://brainglobe.info/developers/index.html).\n\n## License\n\nDistributed under the terms of the [MIT] license,\n\"brainglobe-napari-io\" is free and open source software\n\n## Issues\n\nIf you encounter any problems, please [file an issue] along with a detailed description.\n\n[napari]: https://github.com/napari/napari\n[Cookiecutter]: https://github.com/audreyr/cookiecutter\n[@napari]: https://github.com/napari\n[MIT]: http://opensource.org/licenses/MIT\n[BSD-3]: http://opensource.org/licenses/BSD-3-Clause\n[GNU GPL v3.0]: http://www.gnu.org/licenses/gpl-3.0.txt\n[GNU LGPL v3.0]: http://www.gnu.org/licenses/lgpl-3.0.txt\n[Apache Software License 2.0]: http://www.apache.org/licenses/LICENSE-2.0\n[Mozilla Public License 2.0]: https://www.mozilla.org/media/MPL/2.0/index.txt\n[cookiecutter-napari-plugin]: https://github.com/napari/cookiecutter-napari-plugin\n[file an issue]: https://github.com/brainglobe/brainglobe-napari-io/issues\n[napari]: https://github.com/napari/napari\n[tox]: https://tox.readthedocs.io/en/latest/\n[pip]: https://pypi.org/project/pip/\n[PyPI]: https://pypi.org/\n",
"description_content_type": "text/markdown",
"keywords": null,
"home_page": null,
"download_url": null,
"author": null,
"author_email": "Adam Tyson <[email protected]>",
"maintainer": null,
"maintainer_email": null,
"license": "BSD-3-Clause",
"classifier": [
"Development Status :: 4 - Beta",
"Framework :: napari",
"Intended Audience :: Science/Research",
"Operating System :: OS Independent",
"Programming Language :: Python",
"Programming Language :: Python :: 3",
"Programming Language :: Python :: 3.9",
"Programming Language :: Python :: 3.10",
"Programming Language :: Python :: 3.11",
"Topic :: Scientific/Engineering :: Image Recognition"
],
"requires_dist": [
"brainglobe-atlasapi >=2.0.1",
"brainglobe-space >=1.0.0",
"brainglobe-utils",
"napari",
"tifffile >=2020.8.13",
"numpy",
"pandas",
"pytest ; extra == 'dev'",
"pytest-cov ; extra == 'dev'",
"coverage ; extra == 'dev'",
"tox ; extra == 'dev'",
"black ; extra == 'dev'",
"mypy ; extra == 'dev'",
"pre-commit ; extra == 'dev'",
"ruff ; extra == 'dev'",
"setuptools-scm ; extra == 'dev'"
],
"requires_python": ">=3.9.0",
"requires_external": null,
"project_url": [
"Homepage, https://brainglobe.info",
"Source Code, https://github.com/brainglobe/brainglobe-napari-io",
"Bug Tracker, https://github.com/brainglobe/brainglobe-napari-io/issues",
"Documentation, https://docs.brainglobe.info",
"User Support, https://forum.image.sc/tag/brainglobe",
"Twitter, https://twitter.com/brain_globe"
],
"provides_extra": [
"dev"
],
"provides_dist": null,
"obsoletes_dist": null
},
"npe1_shim": false
}