-
Notifications
You must be signed in to change notification settings - Fork 7
/
PartSeg.json
554 lines (554 loc) · 77.2 KB
/
PartSeg.json
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
{
"name": "PartSeg",
"display_name": "PartSeg",
"visibility": "public",
"icon": "",
"categories": [],
"schema_version": "0.1.0",
"on_activate": null,
"on_deactivate": null,
"contributions": {
"commands": [
{
"id": "PartSeg.load_roi_project",
"title": "Get PartSeg ROI project Reader",
"python_name": "PartSegCore.napari_plugins.load_roi_project:napari_get_reader",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.load_image",
"title": "Get Image Reader",
"python_name": "PartSegCore.napari_plugins.load_image:napari_get_reader",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.write_tiff_image",
"title": "Write tiff Image",
"python_name": "PartSegCore.napari_plugins.save_tiff_layer:napari_write_images",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.write_tiff_labels",
"title": "Write tiff Labels",
"python_name": "PartSegCore.napari_plugins.save_tiff_layer:napari_write_labels",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.load_mask_project",
"title": "Get PartSeg mask project Reader",
"python_name": "PartSegCore.napari_plugins.load_mask_project:napari_get_reader",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.save_mask_roi",
"title": "Write Labels as mask project",
"python_name": "PartSegCore.napari_plugins.save_mask_roi:napari_write_labels",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.load_masked_image",
"title": "Get Reader for image with mask",
"python_name": "PartSegCore.napari_plugins.load_masked_image:napari_get_reader",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.SimpleMeasurement",
"title": "Create Simple Measurement",
"python_name": "PartSeg.plugins.napari_widgets.simple_measurement_widget:SimpleMeasurement",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.ROIAnalysisExtraction",
"title": "Create ROI Analysis Extraction",
"python_name": "PartSeg.plugins.napari_widgets:ROIAnalysisExtraction",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.DoubleThreshold",
"title": "Create Double Threshold",
"python_name": "PartSeg.plugins.napari_widgets:DoubleThreshold",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.NoiseFilter",
"title": "Create Noise Filter",
"python_name": "PartSeg.plugins.napari_widgets:NoiseFilter",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.BorderSmooth",
"title": "Create Border Smooth",
"python_name": "PartSeg.plugins.napari_widgets:BorderSmooth",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.Watershed",
"title": "Create Watershed",
"python_name": "PartSeg.plugins.napari_widgets:Watershed",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.ConnectedComponents",
"title": "Create Connected Components",
"python_name": "PartSeg.plugins.napari_widgets:ConnectedComponents",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.SplitCoreObjects",
"title": "Create Split Core Objects",
"python_name": "PartSeg.plugins.napari_widgets:SplitCoreObjects",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.ROIMaskExtraction",
"title": "Create ROI Mask Extraction",
"python_name": "PartSeg.plugins.napari_widgets:ROIMaskExtraction",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.MaskCreate",
"title": "Create Mask Create",
"python_name": "PartSeg.plugins.napari_widgets:MaskCreate",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.Measurement",
"title": "Create Measurement",
"python_name": "PartSeg.plugins.napari_widgets.measurement_widget:Measurement",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.SearchLabel",
"title": "Create Search Label",
"python_name": "PartSeg.plugins.napari_widgets:SearchLabel",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.PartSegGUILauncher",
"title": "Create Part Seg GUI Launcher",
"python_name": "PartSeg.plugins.napari_widgets:PartSegGUILauncher",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.ImageColormap",
"title": "Create Image Colormap",
"python_name": "PartSeg.plugins.napari_widgets:ImageColormap",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.LabelSelector",
"title": "Create Label Selector",
"python_name": "PartSeg.plugins.napari_widgets:LabelSelector",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.Threshold",
"title": "Create Threshold",
"python_name": "PartSeg.plugins.napari_widgets:Threshold",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.CopyLabels",
"title": "Create Copy Labels",
"python_name": "PartSeg.plugins.napari_widgets:CopyLabelsWidget",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
},
{
"id": "PartSeg.Metadata",
"title": "View Layer Metadata",
"python_name": "PartSeg.plugins.napari_widgets:LayerMetadata",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
}
],
"readers": [
{
"command": "PartSeg.load_roi_project",
"filename_patterns": [
"*.tgz",
"*.tbz2",
"*.gz",
"*.bz2"
],
"accepts_directories": false
},
{
"command": "PartSeg.load_image",
"filename_patterns": [
"*.tif",
"*.tiff",
"*.lsm",
"*.czi",
"*.oib",
"*.oif",
"*.obsep"
],
"accepts_directories": false
},
{
"command": "PartSeg.load_mask_project",
"filename_patterns": [
"*.seg",
"*.tgz"
],
"accepts_directories": false
},
{
"command": "PartSeg.load_masked_image",
"filename_patterns": [
"*.tif",
"*.tiff",
"*.lsm",
"*.czi",
"*.oib",
"*.oif"
],
"accepts_directories": false
}
],
"writers": [
{
"command": "PartSeg.write_tiff_image",
"layer_types": [
"image+"
],
"filename_extensions": [
".tif",
".tiff"
],
"display_name": "Image as tiff"
},
{
"command": "PartSeg.write_tiff_labels",
"layer_types": [
"labels"
],
"filename_extensions": [
".tif",
".tiff"
],
"display_name": "Labels as tiff"
},
{
"command": "PartSeg.save_mask_roi",
"layer_types": [
"labels"
],
"filename_extensions": [
".seg",
".tgz"
],
"display_name": "PartSeg mask roi"
}
],
"widgets": [
{
"command": "PartSeg.SearchLabel",
"display_name": "Search Label",
"autogenerate": false
},
{
"command": "PartSeg.Measurement",
"display_name": "Measurement",
"autogenerate": false
},
{
"command": "PartSeg.SimpleMeasurement",
"display_name": "Simple Measurement",
"autogenerate": false
},
{
"command": "PartSeg.ROIAnalysisExtraction",
"display_name": "ROI Analysis Extraction",
"autogenerate": false
},
{
"command": "PartSeg.ROIMaskExtraction",
"display_name": "ROI Mask Extraction",
"autogenerate": false
},
{
"command": "PartSeg.MaskCreate",
"display_name": "Mask Create",
"autogenerate": false
},
{
"command": "PartSeg.PartSegGUILauncher",
"display_name": "PartSeg GUI Launcher",
"autogenerate": false
},
{
"command": "PartSeg.ImageColormap",
"display_name": "Image Colormap",
"autogenerate": false
},
{
"command": "PartSeg.LabelSelector",
"display_name": "Label Selector",
"autogenerate": false
},
{
"command": "PartSeg.CopyLabels",
"display_name": "Copy Labels",
"autogenerate": false
},
{
"command": "PartSeg.BorderSmooth",
"display_name": "Border Smooth",
"autogenerate": false
},
{
"command": "PartSeg.ConnectedComponents",
"display_name": "Connected Components",
"autogenerate": false
},
{
"command": "PartSeg.NoiseFilter",
"display_name": "Noise Filter",
"autogenerate": false
},
{
"command": "PartSeg.DoubleThreshold",
"display_name": "Double Threshold",
"autogenerate": false
},
{
"command": "PartSeg.SplitCoreObjects",
"display_name": "Split Core Objects",
"autogenerate": false
},
{
"command": "PartSeg.Threshold",
"display_name": "Threshold",
"autogenerate": false
},
{
"command": "PartSeg.Watershed",
"display_name": "Watershed",
"autogenerate": false
},
{
"command": "PartSeg.Metadata",
"display_name": "Layer Metadata",
"autogenerate": false
}
],
"sample_data": null,
"themes": null,
"menus": {},
"submenus": null,
"keybindings": null,
"configuration": []
},
"package_metadata": {
"metadata_version": "2.1",
"name": "PartSeg",
"version": "0.15.4",
"dynamic": null,
"platform": [
"Linux",
"Windows",
"MacOs"
],
"supported_platform": null,
"summary": "PartSeg is python GUI and set of napari plugins for bio imaging analysis especially nucleus analysis,",
"description": "# PartSeg\n\n\n\n[](https://badge.fury.io/py/PartSeg)\n[](https://anaconda.org/conda-forge/partseg)\n[](https://pypi.org/project/partseg)\n[](https://partseg.readthedocs.io/en/latest/?badge=latest)\n[](https://dev.azure.com/PartSeg/PartSeg/_build/latest?definitionId=1&branchName=develop)\n[](https://zenodo.org/badge/latestdoi/166421141)\n[](https://doi.org/10.1186/s12859-021-03984-1)\n[](https://github.com/4DNucleome/PartSeg/blob/master/License.txt)\n[](https://github.com/pre-commit/pre-commit)\n[](https://github.com/psf/black)\n[](https://github.com/charliermarsh/ruff)\n[](https://github.com/4DNucleome/PartSeg/actions/workflows/codeql-analysis.yml)\n[](https://www.codacy.com/gh/4DNucleome/PartSeg/dashboard?utm_source=github.com&utm_medium=referral&utm_content=4DNucleome/PartSeg&utm_campaign=Badge_Grade)\n[](https://codecov.io/gh/4DNucleome/PartSeg)\n[](https://deepsource.io/gh/4DNucleome/PartSeg/?ref=repository-badge)\n\nPartSeg is a GUI and a library for segmentation algorithms. PartSeg also provide napari plugins for IO and labels measurement.\n\nThis application is designed to help biologist with segmentation based on threshold and connected components.\n\n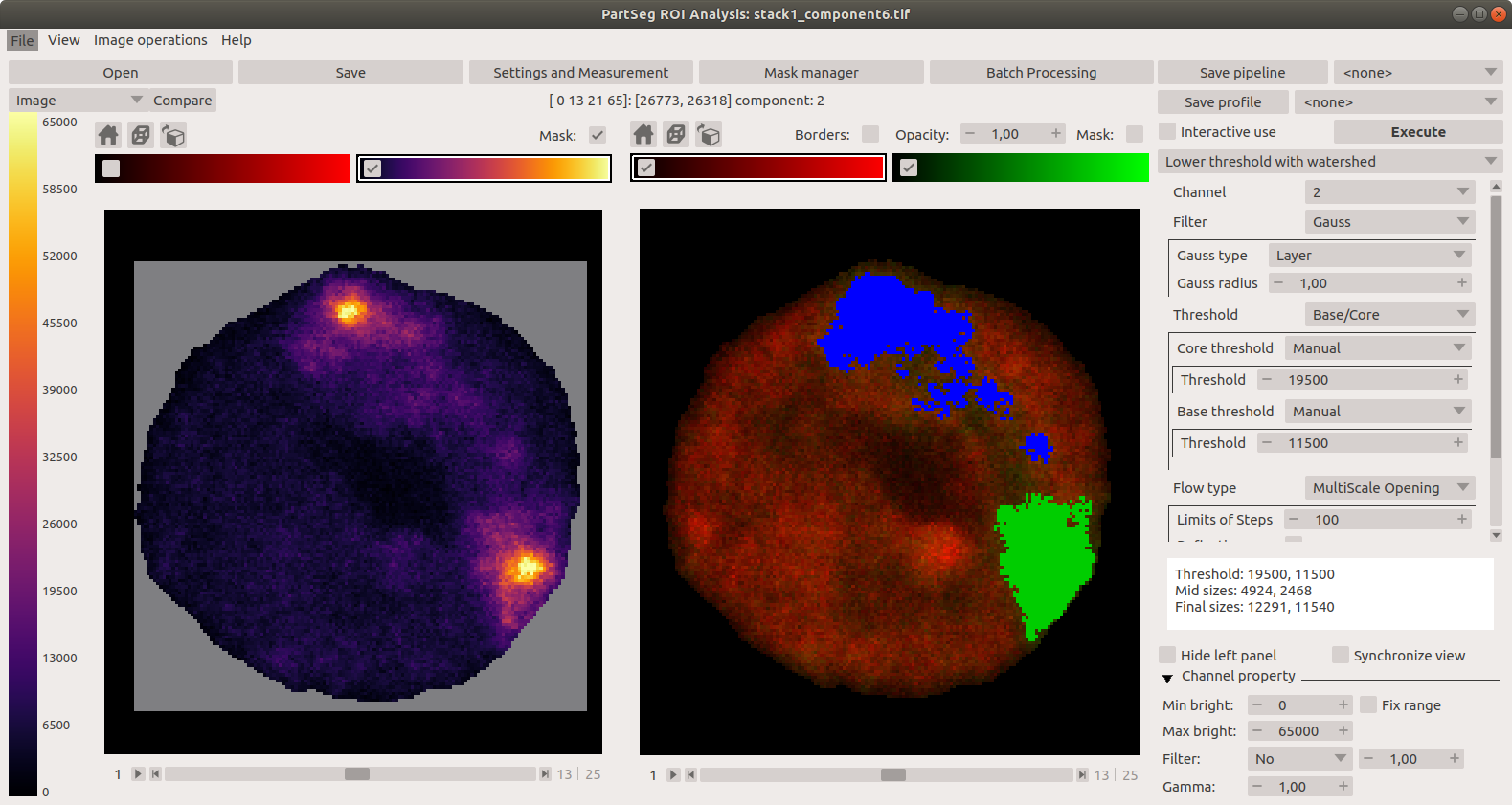\n\n## Tutorials\n\n- Tutorial: **Chromosome 1 (as gui)** [link](https://github.com/4DNucleome/PartSeg/blob/master/tutorials/tutorial-chromosome-1/tutorial-chromosome1_16.md)\n- Data for chromosome 1 tutorial [link](https://4dnucleome.cent.uw.edu.pl/PartSeg/Downloads/PartSeg_samples.zip)\n- Tutorial: **Different neuron types (as library)** [link](https://github.com/4DNucleome/PartSeg/blob/master/tutorials/tutorial_neuron_types/Neuron_types_example.ipynb)\n\n## Installation\n\n- From binaries:\n\n - [Windows](https://github.com/4DNucleome/PartSeg/releases/download/v0.15.4/PartSeg-0.15.4-windows.zip) (build on Windows 10)\n - [Linux](https://github.com/4DNucleome/PartSeg/releases/download/v0.15.4/PartSeg-0.15.4-linux.zip) (build on Ubuntu 20.04)\n - [macOS](https://github.com/4DNucleome/PartSeg/releases/download/v0.15.4/PartSeg-0.15.4-macos.zip) (build on macOS 13)\n - [macOS arm](https://github.com/4DNucleome/PartSeg/releases/download/v0.15.4/PartSeg-0.15.4-macos-arm64.zip) (build on macOS 14)\n There are reported problems with permissions systems on macOS. If you have a problem with starting the application, please try to run it from the terminal.\n\n- With pip:\n\n - From pypi: `pip install PartSeg[all]`\n - From repository: `pip install git+https://github.com/4DNucleome/PartSeg.git`\n\n- With conda:\n\n - `conda install -c conda-forge partseg`\n - `mamba install -c conda-forge partseg` - As mamba is faster than conda\n\n- With napari:\n\n If you do not know how to setup python environment on your system you may use [napari](https://napari.org/) to run PartSeg.\n It is a GUI for scientific image analysis. PartSeg is also a plugin for napari so could be installed from plugin dialog.\n To install napari bundle please download it [napari bundle](https://github.com/napari/napari/releases/latest)\n and follow [installation instructions](https://napari.org/stable/tutorials/fundamentals/installation.html#install-as-a-bundled-app).\n\nInstallation troubleshooting information could be found in wiki: [wiki](https://github.com/4DNucleome/PartSeg/wiki/Instalation-troubleshoot).\nIf this information does not solve problem you can open [issue](https://github.com/4DNucleome/PartSeg/issues).\n\n### Qt 6 support\n\nPartSeg development branch support (and stable since 0.15.0) has experimental Qt6 support. Test are passing but not whole GUI code is covered by tests. Inf you Find any problem please report it.\n\n## Running\n\nIf you downloaded binaries, run the `PartSeg` (or `PartSeg.exe` for Windows) file inside the `PartSeg` folder\n\nIf you installed from repository or from pip, you can run it with `PartSeg` command or `python -m PartSeg`.\nFirst option does not work on Windows.\n\nPartSeg export few commandline options:\n\n- `--no_report` - disable error reporting\n- `--no_dialog` - disable error reporting and error dialog. Use only when running from terminal.\n- `roi` - skip launcher and start *ROI analysis* gui\n- `mask`- skip launcher and start *ROI mask* gui\n\n## napari plugin\n\nPartSeg provides napari plugins for io to allow reading projects format in napari viewer.\n\n## Save Format\n\nSaved projects are tar files compressed with gzip or bz2.\n\nMetadata is saved in data.json file (in json format).\nImages/masks are saved as \\*.npy (numpy array format).\n\n## Interface\n\nLauncher. Choose the program that you will launch:\n\n\n\nMain window of Segmentation Analysis:\n\n\n\nMain window of Segmentation Analysis with view on measurement result:\n\n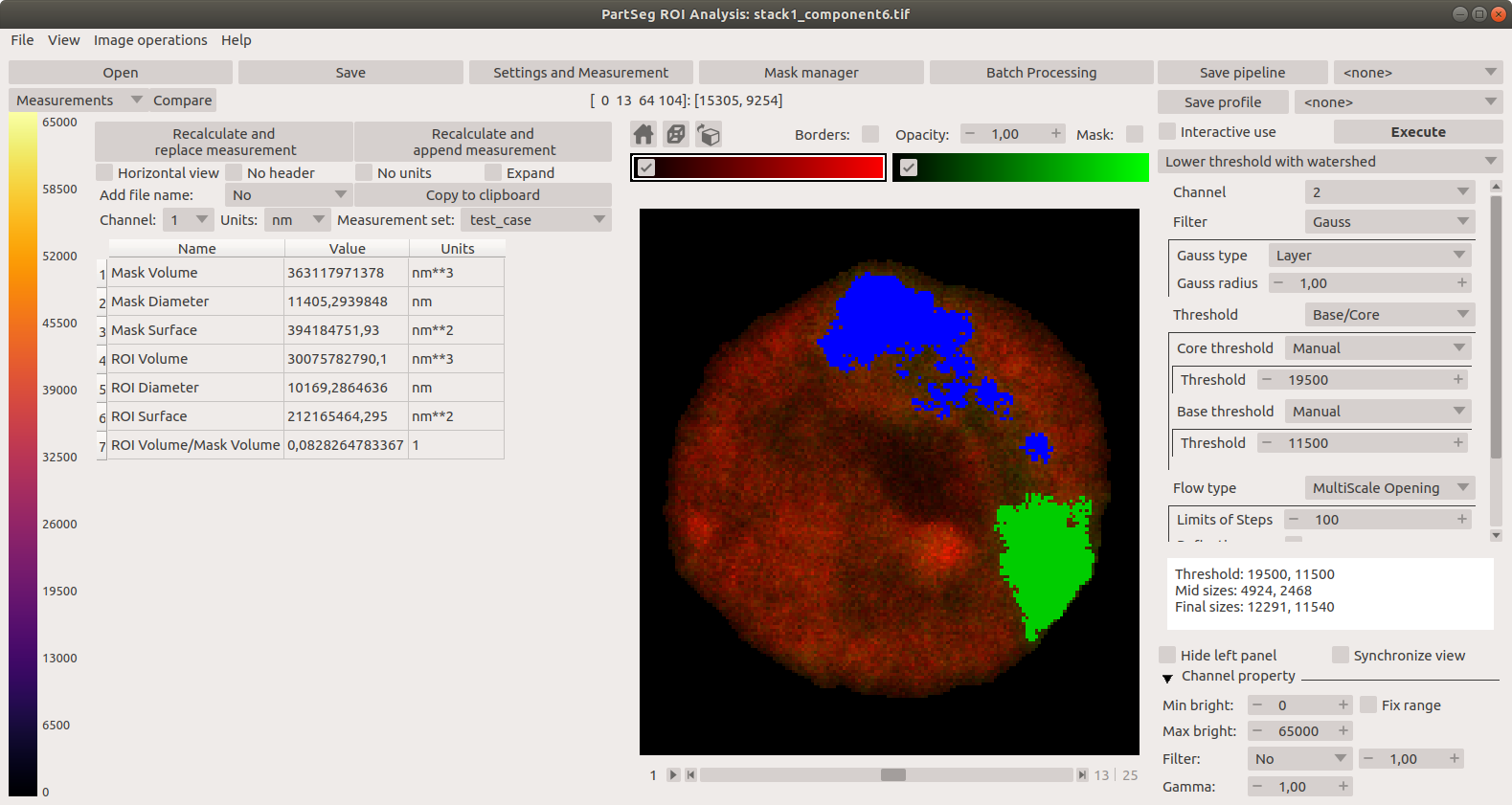\n\nWindow for creating a set of measurements:\n\n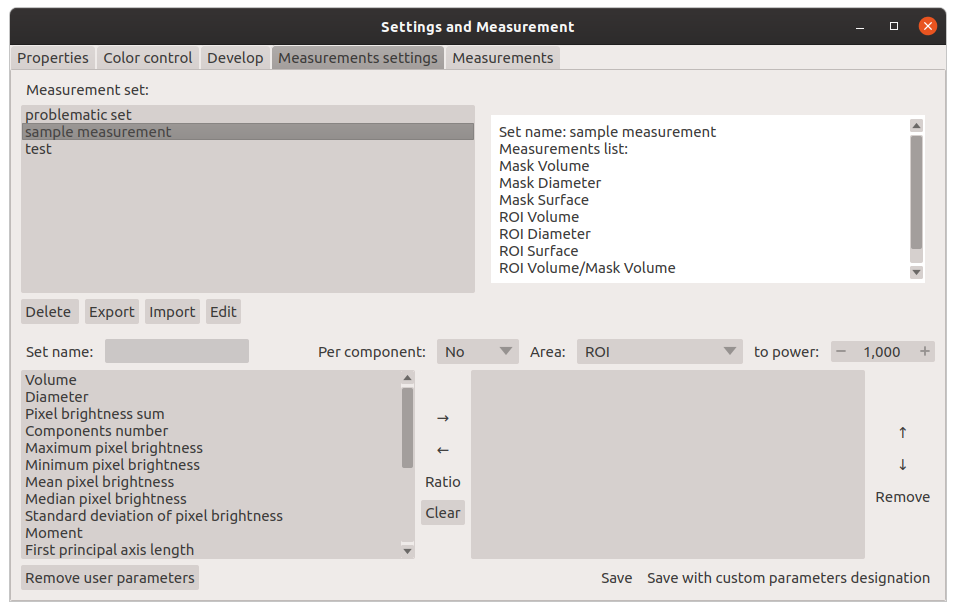\n\nMain window of Mask Segmentation:\n\n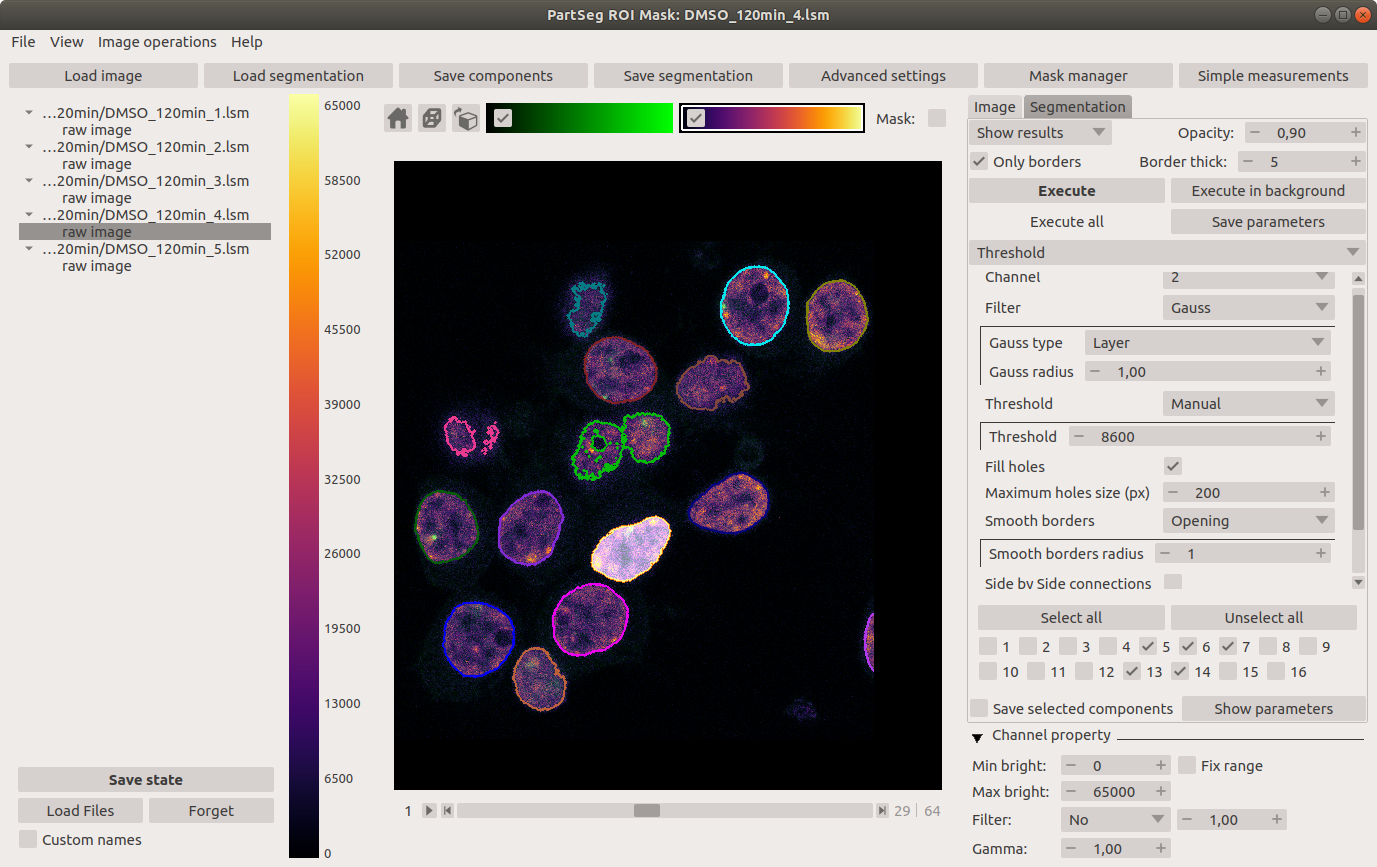\n\n## Laboratory\n\nLaboratory of Functional and Structural Genomics\n[http://4dnucleome.cent.uw.edu.pl/](http://4dnucleome.cent.uw.edu.pl/)\n\n## Cite as\n\nBokota, G., Sroka, J., Basu, S. et al. PartSeg: a tool for quantitative feature extraction\nfrom 3D microscopy images for dummies. BMC Bioinformatics 22, 72 (2021).\n[https://doi.org/10.1186/s12859-021-03984-1](https://doi.org/10.1186/s12859-021-03984-1)\n\n\n## Changelog\n\nAll notable changes to this project will be documented in this file.\n\n### 0.15.4 - 2024-09-27\n\n#### \ud83d\ude80 Features\n\n- Add preview of image metadata ([#1154](https://github.com/4DNucleome/PartSeg/pull/1154))\n- Add option to combine channels using sum and max ([#1159](https://github.com/4DNucleome/PartSeg/pull/1159))\n- Add metadata viewer as napari widget ([#1195](https://github.com/4DNucleome/PartSeg/pull/1195))\n- Read channel colors from `*.czi` metadata ([#1198](https://github.com/4DNucleome/PartSeg/pull/1198))\n- Use image color when add layer to napari ([#1200](https://github.com/4DNucleome/PartSeg/pull/1200))\n\n#### \ud83d\udc1b Bug Fixes\n\n- Fix selection of custom label colors for napari 0.5.0 ([#1138](https://github.com/4DNucleome/PartSeg/pull/1138))\n- Add pint call to enforce initialization of unit registry ([#1146](https://github.com/4DNucleome/PartSeg/pull/1146))\n- Workaround for lack of zsd support in czifile ([#1142](https://github.com/4DNucleome/PartSeg/pull/1142))\n- Fix preparing data for `mahotas.haralick` to avoid overflow problem ([#1150](https://github.com/4DNucleome/PartSeg/pull/1150))\n- Fix `use_convex` type from `int` to `bool` for segmentation algorithms ([#1152](https://github.com/4DNucleome/PartSeg/pull/1152))\n- Prevent propagation of decreasing contrast limits set by user ([#1166](https://github.com/4DNucleome/PartSeg/pull/1166))\n- Prevent error on searching component if there is no component ([#1167](https://github.com/4DNucleome/PartSeg/pull/1167))\n- Fix checking if channel requested by MeasurementProfile exists ([#1165](https://github.com/4DNucleome/PartSeg/pull/1165))\n- Fix trying to access to just deleted measurement profile from edit window. ([#1168](https://github.com/4DNucleome/PartSeg/pull/1168))\n- Fix bug in code for checking for survey file ([#1174](https://github.com/4DNucleome/PartSeg/pull/1174))\n- Fix plugin discovery in bundle to register them in napari viewer ([#1175](https://github.com/4DNucleome/PartSeg/pull/1175))\n- Fix problem with setting range of auto-generated widget ([#1187](https://github.com/4DNucleome/PartSeg/pull/1187))\n- Fix reading channel names from single channel czi files ([#1194](https://github.com/4DNucleome/PartSeg/pull/1194))\n\n#### \ud83d\ude9c Refactor\n\n- Make warnings error in tests ([#1192](https://github.com/4DNucleome/PartSeg/pull/1192))\n- Merge all channel-specific attributes of the Image class ([#1191](https://github.com/4DNucleome/PartSeg/pull/1191))\n\n#### \ud83d\udcda Documentation\n\n- Change homepage URL ([#1139](https://github.com/4DNucleome/PartSeg/pull/1139))\n- Add link for download macOS arm bundle ([#1140](https://github.com/4DNucleome/PartSeg/pull/1140))\n- Add changelog for 0.15.4 release\n- Update changelog ([#1176](https://github.com/4DNucleome/PartSeg/pull/1176))\n\n#### \ud83e\uddea Testing\n\n- \\[Automatic\\] Constraints upgrades: `napari`, `sentry-sdk`, `sympy` ([#1128](https://github.com/4DNucleome/PartSeg/pull/1128))\n- \\[Automatic\\] Constraints upgrades: `mahotas`, `numpy`, `sentry-sdk`, `sympy` ([#1145](https://github.com/4DNucleome/PartSeg/pull/1145))\n- \\[Automatic\\] Constraints upgrades: `numpy`, `tifffile` ([#1163](https://github.com/4DNucleome/PartSeg/pull/1163))\n- \\[Automatic\\] Constraints upgrades: `napari`, `sentry-sdk`, `tifffile` ([#1169](https://github.com/4DNucleome/PartSeg/pull/1169))\n- \\[Automatic\\] Constraints upgrades: `magicgui`, `sentry-sdk` ([#1172](https://github.com/4DNucleome/PartSeg/pull/1172))\n- \\[Automatic\\] Constraints upgrades: `sympy`, `tifffile` ([#1177](https://github.com/4DNucleome/PartSeg/pull/1177))\n- \\[Automatic\\] Constraints upgrades: `imageio`, `napari`, `numpy` ([#1180](https://github.com/4DNucleome/PartSeg/pull/1180))\n- Constraints upgrades: `sentry-sdk` and fix tests ([#1182](https://github.com/4DNucleome/PartSeg/pull/1182))\n- `napari==0.5.3` related fixes, Constraints upgrades: `imageio`, `ipython`, `numpy`, `qtconsole`, `scipy`, `simpleitk`, `tifffile` ([#1183](https://github.com/4DNucleome/PartSeg/pull/1183))\n- \\[Automatic\\] Constraints upgrades: `numpy`, `pydantic` ([#1188](https://github.com/4DNucleome/PartSeg/pull/1188))\n- \\[Automatic\\] Constraints upgrades: `imagecodecs`, `pandas`, `pydantic`, `sentry-sdk`, `sympy`, `tifffile` ([#1190](https://github.com/4DNucleome/PartSeg/pull/1190))\n\n#### \u2699\ufe0f Miscellaneous Tasks\n\n- Speedup tests by use `tox-uv` ([#1141](https://github.com/4DNucleome/PartSeg/pull/1141))\n- Get additional dict from PR branch for checking PR title ([#1144](https://github.com/4DNucleome/PartSeg/pull/1144))\n- Relax numpy constraint ([#1143](https://github.com/4DNucleome/PartSeg/pull/1143))\n- Allow to skip spellchecking PR title ([#1147](https://github.com/4DNucleome/PartSeg/pull/1147))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1149](https://github.com/4DNucleome/PartSeg/pull/1149))\n- Create only archive with version in name on azures pipeline ([#1151](https://github.com/4DNucleome/PartSeg/pull/1151))\n- Fix tests for napari from repository ([#1148](https://github.com/4DNucleome/PartSeg/pull/1148))\n- Use python 3.11 to determine updated packages in PR description ([#1160](https://github.com/4DNucleome/PartSeg/pull/1160))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1164](https://github.com/4DNucleome/PartSeg/pull/1164))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1170](https://github.com/4DNucleome/PartSeg/pull/1170))\n- Disable thumbnail generation in napari layer as it is fragile and not used ([#1171](https://github.com/4DNucleome/PartSeg/pull/1171))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1173](https://github.com/4DNucleome/PartSeg/pull/1173))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1178](https://github.com/4DNucleome/PartSeg/pull/1178))\n- Fix call of logger to properly pass arguments to messages ([#1179](https://github.com/4DNucleome/PartSeg/pull/1179))\n- Fix coverage files upload by enable hidden files upload ([#1186](https://github.com/4DNucleome/PartSeg/pull/1186))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1184](https://github.com/4DNucleome/PartSeg/pull/1184))\n- Use PyQt6 in pre-tests ([#1196](https://github.com/4DNucleome/PartSeg/pull/1196))\n- Add missed code from #1191 ([#1197](https://github.com/4DNucleome/PartSeg/pull/1197))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1189](https://github.com/4DNucleome/PartSeg/pull/1189))\n- Auto add \"skip check PR title\" label in update dependencies PR ([#1199](https://github.com/4DNucleome/PartSeg/pull/1199))\n\n#### Build\n\n- Remove PyOpenGL-accelerate from dependencies because of numpy incompatibility ([#1155](https://github.com/4DNucleome/PartSeg/pull/1155))\n- Update install constraints on numpy and qt packages ([#1157](https://github.com/4DNucleome/PartSeg/pull/1157))\n- Enforce napari 0.5.0 for Qt6 bindings ([#1161](https://github.com/4DNucleome/PartSeg/pull/1161))\n- Require napari>=0.5.0 only for python 3.9+ ([#1162](https://github.com/4DNucleome/PartSeg/pull/1162))\n\n### 0.15.3 - 2024-07-08\n\n#### \ud83d\ude80 Features\n\n- Pydantic 2 compatibility ([#1084](https://github.com/4DNucleome/PartSeg/pull/1084))\n\n#### \ud83d\udc1b Bug Fixes\n\n- Fix rendering icons in colormap preview ([#1040](https://github.com/4DNucleome/PartSeg/pull/1040))\n- Fix test for validation length of message for sentry-sdk 2.0 release ([#1098](https://github.com/4DNucleome/PartSeg/pull/1098))\n- When fix reader check lowercase extension for validate compatibility ([#1097](https://github.com/4DNucleome/PartSeg/pull/1097))\n- Fix napari 0.5.0 compatibility ([#1116](https://github.com/4DNucleome/PartSeg/pull/1116))\n\n#### \ud83d\ude9c Refactor\n\n- Fix Qt flags ([#1041](https://github.com/4DNucleome/PartSeg/pull/1041))\n- Fix qt flags in roi mask code ([#1042](https://github.com/4DNucleome/PartSeg/pull/1042))\n- Fix qt flags in roi analysis ([#1043](https://github.com/4DNucleome/PartSeg/pull/1043))\n- Migrate from setup.cfg to `pyproject.toml` ([#1070](https://github.com/4DNucleome/PartSeg/pull/1070))\n\n#### \ud83d\udcda Documentation\n\n- Allow to use newer release of build docs dependencies ([#1057](https://github.com/4DNucleome/PartSeg/pull/1057))\n\n#### \ud83e\uddea Testing\n\n- \\[Automatic\\] Constraints upgrades: `imagecodecs`, `imageio`, `ipykernel`, `ipython`, `numpy`, `oiffile`, `pandas`, `psygnal`, `pyinstaller`, `qtconsole`, `qtpy`, `sentry-sdk`, `simpleitk`, `superqt`, `tifffile`, `xlsxwriter` ([#1020](https://github.com/4DNucleome/PartSeg/pull/1020))\n- \\[Automatic\\] Constraints upgrades: `h5py`, `imageio`, `ipython`, `numpy`, `packaging`, `pydantic`, `pyinstaller`, `pyqt5`, `scipy`, `sentry-sdk`, `superqt`, `tifffile`, `xlsxwriter` ([#1027](https://github.com/4DNucleome/PartSeg/pull/1027))\n- \\[Automatic\\] Constraints upgrades: `imageio`, `magicgui`, `xlsxwriter` ([#1030](https://github.com/4DNucleome/PartSeg/pull/1030))\n- \\[Automatic\\] Constraints upgrades: `ipykernel`, `pandas`, `qtpy` ([#1032](https://github.com/4DNucleome/PartSeg/pull/1032))\n- \\[Automatic\\] Constraints upgrades: `imageio`, `ipykernel`, `ipython`, `numpy`, `pandas`, `psygnal`, `pygments`, `pyinstaller`, `qtconsole`, `scipy`, `sentry-sdk`, `simpleitk` ([#1035](https://github.com/4DNucleome/PartSeg/pull/1035))\n- \\[Automatic\\] Constraints upgrades: `imagecodecs`, `imageio`, `ipykernel`, `magicgui`, `pandas`, `pyinstaller`, `qtawesome`, `sentry-sdk`, `tifffile` ([#1048](https://github.com/4DNucleome/PartSeg/pull/1048))\n- \\[Automatic\\] Constraints upgrades: `ipykernel`, `numpy`, `pandas`, `partsegcore-compiled-backend`, `pydantic`, `scipy`, `sentry-sdk` ([#1058](https://github.com/4DNucleome/PartSeg/pull/1058))\n- Improve test of PartSegImage ([#1072](https://github.com/4DNucleome/PartSeg/pull/1072))\n- Improve test suite for `PartSegCore` ([#1077](https://github.com/4DNucleome/PartSeg/pull/1077))\n- \\[Automatic\\] Constraints upgrades: `imageio`, `ipykernel`, `local-migrator`, `napari`, `numpy`, `pandas`, `partsegcore-compiled-backend`, `pyinstaller`, `sentry-sdk`, `tifffile`, `vispy`, `xlsxwriter` ([#1063](https://github.com/4DNucleome/PartSeg/pull/1063))\n- \\[Automatic\\] Constraints upgrades: `magicgui`, `packaging`, `psygnal`, `pyinstaller`, `sentry-sdk`, `superqt` ([#1086](https://github.com/4DNucleome/PartSeg/pull/1086))\n- \\[Automatic\\] Constraints upgrades: `psygnal`, `pydantic`, `sentry-sdk`, `vispy` ([#1090](https://github.com/4DNucleome/PartSeg/pull/1090))\n- \\[Automatic\\] Constraints upgrades: `h5py`, `ipykernel`, `mahotas`, `pandas`, `psygnal`, `pydantic`, `pyinstaller`, `qtawesome`, `scipy`, `sentry-sdk`, `superqt` ([#1092](https://github.com/4DNucleome/PartSeg/pull/1092))\n- \\[Automatic\\] Constraints upgrades: `imageio`, `tifffile` ([#1100](https://github.com/4DNucleome/PartSeg/pull/1100))\n- \\[Automatic\\] Constraints upgrades: `pydantic`, `sentry-sdk`, `superqt`, `tifffile` ([#1102](https://github.com/4DNucleome/PartSeg/pull/1102))\n- \\[Automatic\\] Constraints upgrades: `psygnal`, `pygments`, `qtconsole`, `sentry-sdk`, `superqt`, `tifffile` ([#1105](https://github.com/4DNucleome/PartSeg/pull/1105))\n- \\[Automatic\\] Constraints upgrades: `imagecodecs`, `magicgui`, `oiffile`, `openpyxl`, `packaging`, `pydantic`, `pyinstaller`, `requests`, `scipy`, `sentry-sdk`, `superqt`, `sympy`, `tifffile`, `vispy` ([#1107](https://github.com/4DNucleome/PartSeg/pull/1107))\n- \\[Automatic\\] Constraints upgrades: `pydantic` ([#1112](https://github.com/4DNucleome/PartSeg/pull/1112))\n\n#### \u2699\ufe0f Miscellaneous Tasks\n\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1019](https://github.com/4DNucleome/PartSeg/pull/1019))\n- Remove plugin page preview as it is no longer maintained ([#1021](https://github.com/4DNucleome/PartSeg/pull/1021))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1022](https://github.com/4DNucleome/PartSeg/pull/1022))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1026](https://github.com/4DNucleome/PartSeg/pull/1026))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1031](https://github.com/4DNucleome/PartSeg/pull/1031))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1034](https://github.com/4DNucleome/PartSeg/pull/1034))\n- Use new semgrep configuration ([#1039](https://github.com/4DNucleome/PartSeg/pull/1039))\n- Upload raw coverage information ([#1044](https://github.com/4DNucleome/PartSeg/pull/1044))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1036](https://github.com/4DNucleome/PartSeg/pull/1036))\n- Run coverage upload in separate steep ([#1053](https://github.com/4DNucleome/PartSeg/pull/1053))\n- Generate local report in `Tests` workflow and use proper script for fetch report ([#1054](https://github.com/4DNucleome/PartSeg/pull/1054))\n- Move coverage back to main workflow ([#1055](https://github.com/4DNucleome/PartSeg/pull/1055))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1056](https://github.com/4DNucleome/PartSeg/pull/1056))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1059](https://github.com/4DNucleome/PartSeg/pull/1059))\n- Update `actions/upload-artifact` and `actions/download-artifact` from 3 to 4 ([#1062](https://github.com/4DNucleome/PartSeg/pull/1062))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1064](https://github.com/4DNucleome/PartSeg/pull/1064))\n- Group actions update ([#1065](https://github.com/4DNucleome/PartSeg/pull/1065))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1068](https://github.com/4DNucleome/PartSeg/pull/1068))\n- Remove requirement of 2 builds upload to codecov.io ([#1073](https://github.com/4DNucleome/PartSeg/pull/1073))\n- Re add tests to coverage report ([#1074](https://github.com/4DNucleome/PartSeg/pull/1074))\n- Switch from setup.cfg to pyproject.toml in workflows ([#1076](https://github.com/4DNucleome/PartSeg/pull/1076))\n- Fix compiling pyinstaller pre-deps ([#1075](https://github.com/4DNucleome/PartSeg/pull/1075))\n- Add codespell to pre-commit and fix pointed bugs ([#1078](https://github.com/4DNucleome/PartSeg/pull/1078))\n- Add new ruff rules and apply them ([#1079](https://github.com/4DNucleome/PartSeg/pull/1079))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1080](https://github.com/4DNucleome/PartSeg/pull/1080))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1081](https://github.com/4DNucleome/PartSeg/pull/1081))\n- Fix upgrade depenecies workflow ([#1083](https://github.com/4DNucleome/PartSeg/pull/1083))\n- Block using `mpmath==1.4.0a0` and `sentry-sdk` 2.0.0a1/a2 in pre-test ([#1085](https://github.com/4DNucleome/PartSeg/pull/1085))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1089](https://github.com/4DNucleome/PartSeg/pull/1089))\n- Fix jupyter failing test by using constraints ([#1093](https://github.com/4DNucleome/PartSeg/pull/1093))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1091](https://github.com/4DNucleome/PartSeg/pull/1091))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1096](https://github.com/4DNucleome/PartSeg/pull/1096))\n- Add python 3.12 testing ([#1087](https://github.com/4DNucleome/PartSeg/pull/1087))\n- Exclude pyside2 on python 3.11 and 3.12 from testing ([#1099](https://github.com/4DNucleome/PartSeg/pull/1099))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1101](https://github.com/4DNucleome/PartSeg/pull/1101))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1103](https://github.com/4DNucleome/PartSeg/pull/1103))\n- Bump macos runners to macos-13 (both azure and GHA) ([#1113](https://github.com/4DNucleome/PartSeg/pull/1113))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1108](https://github.com/4DNucleome/PartSeg/pull/1108))\n- Remove pyqt5 from constraints ([#1118](https://github.com/4DNucleome/PartSeg/pull/1118))\n- Add workflow for releases from GHA ([#1117](https://github.com/4DNucleome/PartSeg/pull/1117))\n- Add actionlint to CI to early prevent bug in github workflows ([#1119](https://github.com/4DNucleome/PartSeg/pull/1119))\n- Fix release workflow, by update permissions\n- Check if release notes are properly created ([#1122](https://github.com/4DNucleome/PartSeg/pull/1122))\n- Proper use enum in checking new version ([#1123](https://github.com/4DNucleome/PartSeg/pull/1123))\n- Refactor and simplify menu bar creation, add workaround for macOS numpy problem ([#1124](https://github.com/4DNucleome/PartSeg/pull/1124))\n- Simplify release workflow ([#1126](https://github.com/4DNucleome/PartSeg/pull/1126))\n- Fix `make_release.yml` to proper detect release, attempt 3 ([#1127](https://github.com/4DNucleome/PartSeg/pull/1127))\n\n#### \ud83d\udee1\ufe0f Security\n\n- *(deps)* Bump actions/checkout from 3 to 4 ([#1029](https://github.com/4DNucleome/PartSeg/pull/1029))\n- *(deps)* Bump conda-incubator/setup-miniconda from 2 to 3 ([#1038](https://github.com/4DNucleome/PartSeg/pull/1038))\n- *(deps)* Bump aganders3/headless-gui from 1 to 2 ([#1047](https://github.com/4DNucleome/PartSeg/pull/1047))\n- *(deps)* Bump actions/checkout from 3 to 4 ([#1045](https://github.com/4DNucleome/PartSeg/pull/1045))\n- *(deps)* Bump hynek/build-and-inspect-python-package from 1 to 2 ([#1050](https://github.com/4DNucleome/PartSeg/pull/1050))\n- *(deps)* Bump actions/setup-python from 4 to 5 ([#1046](https://github.com/4DNucleome/PartSeg/pull/1046))\n- *(deps)* Bump github/codeql-action from 2 to 3 ([#1051](https://github.com/4DNucleome/PartSeg/pull/1051))\n- *(deps)* Bump peter-evans/create-pull-request from 5 to 6 ([#1067](https://github.com/4DNucleome/PartSeg/pull/1067))\n- *(deps)* Bump codecov/codecov-action from 3 to 4 ([#1066](https://github.com/4DNucleome/PartSeg/pull/1066))\n\n#### Build\n\n- Fix not bundling `Font Awesome 6 Free-Solid-900.otf` file to executable ([#1114](https://github.com/4DNucleome/PartSeg/pull/1114))\n- Update readme and release to point to GitHub releases ([#1115](https://github.com/4DNucleome/PartSeg/pull/1115))\n- Do not create archive twice when create bundle ([#1120](https://github.com/4DNucleome/PartSeg/pull/1120))\n- Enable macOS-arm bundle builds ([#1121](https://github.com/4DNucleome/PartSeg/pull/1121))\n\n### 0.15.2 - 2023-08-28\n\n#### \ud83d\udc1b Bug Fixes\n\n- Fix range threshold selection of algorithms ([#1009](https://github.com/4DNucleome/PartSeg/pull/1009))\n- When run batch check if file extension is supported by loader ([#1016](https://github.com/4DNucleome/PartSeg/pull/1016))\n- Do not allow to select and render corrupted batch plans ([#1015](https://github.com/4DNucleome/PartSeg/pull/1015))\n\n#### \ud83e\uddea Testing\n\n- \\[Automatic\\] Constraints upgrades: `imagecodecs`, `ipykernel`, `magicgui`, `psygnal`, `scipy`, `superqt`, `tifffile` ([#1011](https://github.com/4DNucleome/PartSeg/pull/1011))\n- \\[Automatic\\] Constraints upgrades: `imageio`, `pyinstaller`, `tifffile` ([#1018](https://github.com/4DNucleome/PartSeg/pull/1018))\n\n#### \u2699\ufe0f Miscellaneous Tasks\n\n- Use faster version of black ([#1010](https://github.com/4DNucleome/PartSeg/pull/1010))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1013](https://github.com/4DNucleome/PartSeg/pull/1013))\n\n### 0.15.1 - 2023-08-08\n\n#### \ud83d\ude80 Features\n\n- Allow to save multiple napari image layers to single tiff file ([#1000](https://github.com/4DNucleome/PartSeg/pull/1000))\n- Add option to export batch project with data ([#996](https://github.com/4DNucleome/PartSeg/pull/996))\n\n#### \ud83d\udc1b Bug Fixes\n\n- Fix possible problem of double registration napari plugin in PartSeg bundle ([#974](https://github.com/4DNucleome/PartSeg/pull/974))\n- Bump OS versions for part of testing workflows. ([#977](https://github.com/4DNucleome/PartSeg/pull/977))\n- Bump os version for main tests workflow. ([#979](https://github.com/4DNucleome/PartSeg/pull/979))\n- Ensure that the module `PartSegCore.channel_class` is present in bundle ([#980](https://github.com/4DNucleome/PartSeg/pull/980))\n- Lower npe2 schema version to work with older napari version ([#981](https://github.com/4DNucleome/PartSeg/pull/981))\n- Generate test report per platform ([#978](https://github.com/4DNucleome/PartSeg/pull/978))\n- Importing plugins in bundle keeping proper module names ([#983](https://github.com/4DNucleome/PartSeg/pull/983))\n- Fix napari repo workflow ([#985](https://github.com/4DNucleome/PartSeg/pull/985))\n- Fix bug in read tiff files with double `Q` in axes but one related to dummy dimension ([#992](https://github.com/4DNucleome/PartSeg/pull/992))\n- Fix bug that lead to corrupted state when saving calculation plan to excel file ([#995](https://github.com/4DNucleome/PartSeg/pull/995))\n- Enable python 3.11 test on CI, fix minor errors ([#869](https://github.com/4DNucleome/PartSeg/pull/869))\n\n#### \ud83e\uddea Testing\n\n- \\[Automatic\\] Constraints upgrades: `imageio`, `ipython`, `psygnal`, `scipy`, `sentry-sdk` ([#975](https://github.com/4DNucleome/PartSeg/pull/975))\n- \\[Automatic\\] Constraints upgrades: `h5py`, `imagecodecs`, `imageio`, `ipykernel`, `napari`, `numpy`, `pandas`, `pydantic`, `pyinstaller`, `scipy`, `sentry-sdk`, `tifffile`, `vispy` ([#986](https://github.com/4DNucleome/PartSeg/pull/986))\n- \\[Automatic\\] Constraints upgrades: `imagecodecs`, `sentry-sdk`, `tifffile` ([#997](https://github.com/4DNucleome/PartSeg/pull/997))\n- \\[Automatic\\] Constraints upgrades: `ipykernel`, `pydantic` ([#1002](https://github.com/4DNucleome/PartSeg/pull/1002))\n- \\[Automatic\\] Constraints upgrades: `numpy`, `pygments`, `sentry-sdk`, `superqt` ([#1007](https://github.com/4DNucleome/PartSeg/pull/1007))\n\n#### \u2699\ufe0f Miscellaneous Tasks\n\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#973](https://github.com/4DNucleome/PartSeg/pull/973))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#982](https://github.com/4DNucleome/PartSeg/pull/982))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#987](https://github.com/4DNucleome/PartSeg/pull/987))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#988](https://github.com/4DNucleome/PartSeg/pull/988))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#991](https://github.com/4DNucleome/PartSeg/pull/991))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#998](https://github.com/4DNucleome/PartSeg/pull/998))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1004](https://github.com/4DNucleome/PartSeg/pull/1004))\n- Change markdown linter from pre-commit to mdformat ([#1006](https://github.com/4DNucleome/PartSeg/pull/1006))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#1008](https://github.com/4DNucleome/PartSeg/pull/1008))\n\n### 0.15.0 - 2023-05-30\n\n#### \ud83d\ude80 Features\n\n- Add `PARTSEG_SENTRY_URL` env variable support and basic documentation about error reporting ([#802](https://github.com/4DNucleome/PartSeg/pull/802))\n- Allow to see underlying exception when show warning caused by exception ([#829](https://github.com/4DNucleome/PartSeg/pull/829))\n- Add voxel size measurement and allow to overwrite voxel size in batch ([#853](https://github.com/4DNucleome/PartSeg/pull/853))\n- Add alpha support for Qt6 ([#866](https://github.com/4DNucleome/PartSeg/pull/866))\n- Add option to create projection alongside z-axis ([#919](https://github.com/4DNucleome/PartSeg/pull/919))\n- Add napari image custom representation for better error report via sentry ([#861](https://github.com/4DNucleome/PartSeg/pull/861))\n- Add import and export operation for labels and colormaps ([#936](https://github.com/4DNucleome/PartSeg/pull/936))\n- Implement napari widgets for colormap and labels control ([#935](https://github.com/4DNucleome/PartSeg/pull/935))\n- Add forget all button to multiple files widget ([#942](https://github.com/4DNucleome/PartSeg/pull/942))\n- Do not abort processing whole mask segmentation project during exception on single component ([#943](https://github.com/4DNucleome/PartSeg/pull/943))\n- Add distance based watersheed to flow methods ([#915](https://github.com/4DNucleome/PartSeg/pull/915))\n- Add napari widgets for all group of algorithms ([#958](https://github.com/4DNucleome/PartSeg/pull/958))\n- Add napari widget to copy labels along z-axis ([#968](https://github.com/4DNucleome/PartSeg/pull/968))\n\n#### \ud83d\udc1b Bug Fixes\n\n- Print all exceptions instead of the latest one in exception dialog ([#799](https://github.com/4DNucleome/PartSeg/pull/799))\n- Fix ROIExtractionResult `__str__`and `__repr__` to use `ROIExtractionResult` not `SegmentationResult` ([#810](https://github.com/4DNucleome/PartSeg/pull/810))\n- Fix code to address changes in napari repository ([#817](https://github.com/4DNucleome/PartSeg/pull/817))\n- Fix problem with resize of multiline widgets ([#832](https://github.com/4DNucleome/PartSeg/pull/832))\n- Fix tox configuration to run all required tests ([#840](https://github.com/4DNucleome/PartSeg/pull/840))\n- Fix MSO `step_limit` description in GUI ([#843](https://github.com/4DNucleome/PartSeg/pull/843))\n- Fix `redefined-while-unused`import code for python 3.9.7 ([#844](https://github.com/4DNucleome/PartSeg/pull/844))\n- Fix warnings reported by Deepsource ([#846](https://github.com/4DNucleome/PartSeg/pull/846))\n- Ensure that \"ROI\" layer is in proper place for proper visualization ([#856](https://github.com/4DNucleome/PartSeg/pull/856))\n- Fix tests of napari widgets ([#862](https://github.com/4DNucleome/PartSeg/pull/862))\n- Fix build of bundle for a new psygnal release ([#863](https://github.com/4DNucleome/PartSeg/pull/863))\n- Fix minimal requirements pipeline ([#877](https://github.com/4DNucleome/PartSeg/pull/877))\n- Update pyinstaller configuration ([#926](https://github.com/4DNucleome/PartSeg/pull/926))\n- Use text icon, not pixmap icon in colormap and labels list ([#938](https://github.com/4DNucleome/PartSeg/pull/938))\n- Resolve warnings when testing custom save dialog. ([#941](https://github.com/4DNucleome/PartSeg/pull/941))\n- Add padding zeros for component num when load Mask seg file to ROI GUI ([#944](https://github.com/4DNucleome/PartSeg/pull/944))\n- Proper calculate bounds for watershed napari widget ([#969](https://github.com/4DNucleome/PartSeg/pull/969))\n- Fix bug in the wrong order of axis saved in napari contribution ([#972](https://github.com/4DNucleome/PartSeg/pull/972))\n\n#### \ud83d\ude9c Refactor\n\n- Simplify and refactor github workflows. ([#864](https://github.com/4DNucleome/PartSeg/pull/864))\n- Better load Mask project in Roi Analysis ([#921](https://github.com/4DNucleome/PartSeg/pull/921))\n- Use more descriptive names in `pylint: disable` ([#922](https://github.com/4DNucleome/PartSeg/pull/922))\n- Remove `pkg_resources` usage as it is deprecated ([#967](https://github.com/4DNucleome/PartSeg/pull/967))\n- Convert napari plugin to npe2 ([#966](https://github.com/4DNucleome/PartSeg/pull/966))\n\n#### \ud83d\udcda Documentation\n\n- Update README and project metadata ([#805](https://github.com/4DNucleome/PartSeg/pull/805))\n- Create release notes for PartSeg 0.15.0 ([#971](https://github.com/4DNucleome/PartSeg/pull/971))\n\n#### \ud83c\udfa8 Styling\n\n- Change default theme to dark, remove blinking windows on startup. ([#809](https://github.com/4DNucleome/PartSeg/pull/809))\n\n#### \ud83e\uddea Testing\n\n- \\[Automatic\\] Dependency upgrades: `packaging`, `pyinstaller`, `pyopengl-accelerate`, `tifffile`, `xlsxwriter` ([#932](https://github.com/4DNucleome/PartSeg/pull/932))\n- \\[Automatic\\] Constraints upgrades: `fonticon-fontawesome6`, `imageio`, `numpy`, `partsegcore-compiled-backend`, `pygments`, `sentry-sdk` ([#937](https://github.com/4DNucleome/PartSeg/pull/937))\n- \\[Automatic\\] Constraints upgrades: `imageio`, `ipython`, `pandas`, `requests`, `sentry-sdk` ([#948](https://github.com/4DNucleome/PartSeg/pull/948))\n- \\[Automatic\\] Constraints upgrades: `ipython`, `nme`, `qtconsole`, `requests`, `sentry-sdk` ([#955](https://github.com/4DNucleome/PartSeg/pull/955))\n- \\[Automatic\\] Constraints upgrades: `ipykernel`, `local-migrator`, `pyinstaller`, `sentry-sdk`, `sympy` ([#957](https://github.com/4DNucleome/PartSeg/pull/957))\n- \\[Automatic\\] Constraints upgrades: `sentry-sdk`, `xlsxwriter` ([#959](https://github.com/4DNucleome/PartSeg/pull/959))\n- \\[Automatic\\] Constraints upgrades: `requests` ([#961](https://github.com/4DNucleome/PartSeg/pull/961))\n- \\[Automatic\\] Constraints upgrades: `imageio`, `pandas`, `pydantic`, `pyopengl-accelerate`, `sentry-sdk`, `xlsxwriter` ([#970](https://github.com/4DNucleome/PartSeg/pull/970))\n\n#### \u2699\ufe0f Miscellaneous Tasks\n\n- Improve ruff configuration, remove isort ([#815](https://github.com/4DNucleome/PartSeg/pull/815))\n- Use `fail_on_no_env` feature from `tox-gh-actions` ([#842](https://github.com/4DNucleome/PartSeg/pull/842))\n- Add python 3.11 to list of supported versions ([#867](https://github.com/4DNucleome/PartSeg/pull/867))\n- Disable python 3.11 test because of timeout ([#870](https://github.com/4DNucleome/PartSeg/pull/870))\n- Bump ruff to 0.0.218, remove flake8 from pre-commit ([#880](https://github.com/4DNucleome/PartSeg/pull/880))\n- Replace GabrielBB/xvfb-action@v1 by aganders3/headless-gui, part 2 ([#887](https://github.com/4DNucleome/PartSeg/pull/887))\n- Better minimal requirements test ([#888](https://github.com/4DNucleome/PartSeg/pull/888))\n- Improve regexp for proper generate list of packages in update report ([#894](https://github.com/4DNucleome/PartSeg/pull/894))\n- Add check for PR title ([#933](https://github.com/4DNucleome/PartSeg/pull/933))\n- Update codecov configuration to wait on two reports before post information ([#934](https://github.com/4DNucleome/PartSeg/pull/934))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#945](https://github.com/4DNucleome/PartSeg/pull/945))\n- Migrate from `nme` to `local_migrator` ([#951](https://github.com/4DNucleome/PartSeg/pull/951))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#956](https://github.com/4DNucleome/PartSeg/pull/956))\n- \\[pre-commit.ci\\] pre-commit autoupdate ([#964](https://github.com/4DNucleome/PartSeg/pull/964))\n\n#### \ud83d\udee1\ufe0f Security\n\n- *(deps)* Bump peter-evans/create-pull-request from 4 to 5 ([#928](https://github.com/4DNucleome/PartSeg/pull/928))\n\n#### Bugfix\n\n- Fix bug with generation of form for model with hidden field ([#920](https://github.com/4DNucleome/PartSeg/pull/920))\n\n#### Dep\n\n- \\[Automatic\\] Dependency upgrades ([#824](https://github.com/4DNucleome/PartSeg/pull/824))\n- \\[Automatic\\] Dependency upgrades ([#828](https://github.com/4DNucleome/PartSeg/pull/828))\n- \\[Automatic\\] Dependency upgrades: `ipykernel`, `packaging` ([#838](https://github.com/4DNucleome/PartSeg/pull/838))\n- \\[Automatic\\] Dependency upgrades: `imageio`, `ipykernel`, `napari`, `numpy`, `sentry` ([#850](https://github.com/4DNucleome/PartSeg/pull/850))\n- \\[Automatic\\] Dependency upgrades: `imagecodecs`, `ipykernel`, `numpy`, `psygnal` ([#859](https://github.com/4DNucleome/PartSeg/pull/859))\n- \\[Automatic\\] Dependency upgrades: `pydantic`, `pygments`, `xlsxwriter` ([#874](https://github.com/4DNucleome/PartSeg/pull/874))\n- \\[Automatic\\] Dependency upgrades: `imageio`, `packaging`, `scipy`, `xlsxwriter` ([#878](https://github.com/4DNucleome/PartSeg/pull/878))\n- \\[Automatic\\] Dependency upgrades: `ipykernel`, `requests`, `sentry`, `xlsxwriter` ([#884](https://github.com/4DNucleome/PartSeg/pull/884))\n- \\[Automatic\\] Dependency upgrades: `h5py`, `imagecodecs`, `imageio`, `ipykernel`, `pandas`, `sentry`, `tifffile` ([#889](https://github.com/4DNucleome/PartSeg/pull/889))\n- \\[Automatic\\] Dependency upgrades: `ipython`, `pyqt5` ([#893](https://github.com/4DNucleome/PartSeg/pull/893))\n- \\[Automatic\\] Dependency upgrades: `imageio`, `ipykernel`, `ipython`, `numpy`, `openpyxl`, `psygnal`, `pydantic`, `pyinstaller`, `pyqt5`, `scipy`, `sentry-sdk`, `tifffile`, `xlsxwriter` ([#897](https://github.com/4DNucleome/PartSeg/pull/897))\n- \\[Automatic\\] Dependency upgrades: `imageio`, `psygnal` ([#905](https://github.com/4DNucleome/PartSeg/pull/905))\n- \\[Automatic\\] Dependency upgrades: `ipython`, `magicgui`, `scipy`, `sentry-sdk`, `tifffile` ([#906](https://github.com/4DNucleome/PartSeg/pull/906))\n- \\[Automatic\\] Dependency upgrades: `imagecodecs`, `imageio`, `ipykernel`, `openpyxl`, `pydantic`, `pyinstaller`, `qtawesome`, `qtconsole`, `sentry-sdk`, `tifffile`, `xlsxwriter` ([#908](https://github.com/4DNucleome/PartSeg/pull/908))\n- \\[Automatic\\] Dependency upgrades: `imageio`, `ipykernel`, `ipython`, `pandas`, `psygnal`, `pydantic`, `pygments`, `pyinstaller`, `qtpy`, `sentry-sdk`, `tifffile` ([#917](https://github.com/4DNucleome/PartSeg/pull/917))\n\n### 0.14.6 - 2022-11-13\n\n#### \ud83d\udc1b Bug Fixes\n\n- Fix bug when loading already created project causing hide of ROI layer ([#787](https://github.com/4DNucleome/PartSeg/pull/787))\n\n### 0.14.5 - 2022-11-09\n\n#### \ud83d\ude80 Features\n\n- Add option for ensure type in EventedDict and use it to validate profiles structures ([#776](https://github.com/4DNucleome/PartSeg/pull/776))\n- Add option to create issue from error report dialog ([#782](https://github.com/4DNucleome/PartSeg/pull/782))\n- Add option for multiline field in algorithm parameters ([#766](https://github.com/4DNucleome/PartSeg/pull/766))\n\n#### \ud83d\udc1b Bug Fixes\n\n- Fix scalebar color ([#774](https://github.com/4DNucleome/PartSeg/pull/774))\n- Fix bug when saving segmentation parameters in mask analysis ([#781](https://github.com/4DNucleome/PartSeg/pull/781))\n- Fix multiple error related to loading new file in interactive mode ([#784](https://github.com/4DNucleome/PartSeg/pull/784))\n\n#### \ud83d\ude9c Refactor\n\n- Optimize CLI actions ([#772](https://github.com/4DNucleome/PartSeg/pull/772))\n- Clean warnings about threshold methods ([#783](https://github.com/4DNucleome/PartSeg/pull/783))\n\n#### Build\n\n- *(deps)* Bump chanzuckerberg/napari-hub-preview-action from 0.1.5 to 0.1.6 ([#775](https://github.com/4DNucleome/PartSeg/pull/775))\n\n### 0.14.4 - 2022-10-24\n\n#### \ud83d\ude80 Features\n\n- Load alternatives labeling when open PartSeg projects in napari ([#731](https://github.com/4DNucleome/PartSeg/pull/731))\n- Add option to toggle scale bar ([#733](https://github.com/4DNucleome/PartSeg/pull/733))\n- Allow customize settings directory using the `PARTSEG_SETTINGS_DIR` environment variable ([#751](https://github.com/4DNucleome/PartSeg/pull/751))\n- Separate recent algorithms from general application settings ([#752](https://github.com/4DNucleome/PartSeg/pull/752))\n- Add multiple otsu as threshold method with selection range of components ([#710](https://github.com/4DNucleome/PartSeg/pull/710))\n- Add function to load components from Mask Segmentation with background in ROI Analysis ([#768](https://github.com/4DNucleome/PartSeg/pull/768))\n\n#### \ud83d\udc1b Bug Fixes\n\n- Fix typos\n- Fix `get_theme` calls to prepare for napari 0.4.17 ([#729](https://github.com/4DNucleome/PartSeg/pull/729))\n- Fix saving pipeline from GUI ([#756](https://github.com/4DNucleome/PartSeg/pull/756))\n- Fix profile export/import dialogs ([#761](https://github.com/4DNucleome/PartSeg/pull/761))\n- Enable compare button if ROI is available ([#765](https://github.com/4DNucleome/PartSeg/pull/765))\n- Fix bug in cut with roi to do not make black artifacts ([#767](https://github.com/4DNucleome/PartSeg/pull/767))\n\n#### \ud83e\uddea Testing\n\n- Add new build and inspect wheel action ([#747](https://github.com/4DNucleome/PartSeg/pull/747))\n\n#### \u2699\ufe0f Miscellaneous Tasks\n\n- Prepare pyinstaller configuration for napari 0.4.17 ([#748](https://github.com/4DNucleome/PartSeg/pull/748))\n- Add ruff linter ([#754](https://github.com/4DNucleome/PartSeg/pull/754))\n\n#### Bugfix\n\n- Fix sentry tests ([#742](https://github.com/4DNucleome/PartSeg/pull/742))\n- Fix reporting error in load settings from drive ([#725](https://github.com/4DNucleome/PartSeg/pull/725))\n\n#### Build\n\n- *(deps)* Bump actions/checkout from 2 to 3 ([#716](https://github.com/4DNucleome/PartSeg/pull/716))\n- *(deps)* Bump actions/download-artifact from 1 to 3 ([#709](https://github.com/4DNucleome/PartSeg/pull/709))\n\n### 0.14.3 - 2022-08-18\n\n#### \ud83d\udc1b Bug Fixes\n\n- Delay setting image if an algorithm is still running ([#627](https://github.com/4DNucleome/PartSeg/pull/627))\n- Wrong error report when no component is found in restartable segmentation algorithm. ([#633](https://github.com/4DNucleome/PartSeg/pull/633))\n- Fix process of build documentation ([#653](https://github.com/4DNucleome/PartSeg/pull/653))\n\n#### \ud83d\ude9c Refactor\n\n- Clean potential vulnerabilities ([#630](https://github.com/4DNucleome/PartSeg/pull/630))\n\n#### \ud83e\uddea Testing\n\n- Add more tests for common GUI elements ([#622](https://github.com/4DNucleome/PartSeg/pull/622))\n- Report coverage per package. ([#639](https://github.com/4DNucleome/PartSeg/pull/639))\n- Update conda environment to not use PyQt5 in test ([#646](https://github.com/4DNucleome/PartSeg/pull/646))\n- Add tests files to calculate coverage ([#655](https://github.com/4DNucleome/PartSeg/pull/655))\n\n#### Build\n\n- *(deps)* Bump qtpy from 2.0.1 to 2.1.0 in /requirements ([#613](https://github.com/4DNucleome/PartSeg/pull/613))\n- *(deps)* Bump pyinstaller from 5.0.1 to 5.1 in /requirements ([#629](https://github.com/4DNucleome/PartSeg/pull/629))\n- *(deps)* Bump tifffile from 2022.4.28 to 2022.5.4 in /requirements ([#619](https://github.com/4DNucleome/PartSeg/pull/619))\n- *(deps)* Bump codecov/codecov-action from 1 to 3 ([#637](https://github.com/4DNucleome/PartSeg/pull/637))\n- *(deps)* Bump requests from 2.27.1 to 2.28.0 in /requirements ([#647](https://github.com/4DNucleome/PartSeg/pull/647))\n- *(deps)* Bump actions/setup-python from 3 to 4 ([#648](https://github.com/4DNucleome/PartSeg/pull/648))\n- *(deps)* Bump pyqt5 from 5.15.6 to 5.15.7 in /requirements ([#652](https://github.com/4DNucleome/PartSeg/pull/652))\n- *(deps)* Bump sentry-sdk from 1.5.12 to 1.6.0 in /requirements ([#659](https://github.com/4DNucleome/PartSeg/pull/659))\n- *(deps)* Bump numpy from 1.22.4 to 1.23.0 in /requirements ([#660](https://github.com/4DNucleome/PartSeg/pull/660))\n- *(deps)* Bump lxml from 4.9.0 to 4.9.1 in /requirements ([#665](https://github.com/4DNucleome/PartSeg/pull/665))\n- *(deps)* Bump mahotas from 1.4.12 to 1.4.13 in /requirements ([#662](https://github.com/4DNucleome/PartSeg/pull/662))\n- *(deps)* Bump pyinstaller from 5.1 to 5.2 in /requirements ([#667](https://github.com/4DNucleome/PartSeg/pull/667))\n\n### 0.14.2 - 2022-05-05\n\n#### \ud83d\udc1b Bug Fixes\n\n- Fix bug in save label colors between sessions ([#610](https://github.com/4DNucleome/PartSeg/pull/610))\n- Register PartSeg plugins before start napari widgets. ([#611](https://github.com/4DNucleome/PartSeg/pull/611))\n- Mouse interaction with components work again after highlight. ([#620](https://github.com/4DNucleome/PartSeg/pull/620))\n\n#### \ud83d\ude9c Refactor\n\n- Limit test run ([#603](https://github.com/4DNucleome/PartSeg/pull/603))\n- Filter and solve warnings in tests ([#607](https://github.com/4DNucleome/PartSeg/pull/607))\n- Use QAbstractSpinBox.AdaptiveDecimalStepType in SpinBox instead of hardcoded bounds ([#616](https://github.com/4DNucleome/PartSeg/pull/616))\n- Clean and test `PartSeg.common_gui.universal_gui_part` ([#617](https://github.com/4DNucleome/PartSeg/pull/617))\n\n#### \ud83d\udcda Documentation\n\n- Update changelog ([#621](https://github.com/4DNucleome/PartSeg/pull/621))\n\n#### \ud83e\uddea Testing\n\n- Speedup test by setup cache for pip ([#604](https://github.com/4DNucleome/PartSeg/pull/604))\n- Setup cache for azure pipelines workflows ([#606](https://github.com/4DNucleome/PartSeg/pull/606))\n\n#### Build\n\n- *(deps)* Bump sentry-sdk from 1.5.10 to 1.5.11 in /requirements ([#615](https://github.com/4DNucleome/PartSeg/pull/615))\n\n### 0.14.1 - 2022-04-27\n\n#### \ud83d\ude80 Features\n\n- Use pygments for coloring code in exception window ([#591](https://github.com/4DNucleome/PartSeg/pull/591))\n- Add option to calculate Measurement per Mask component ([#590](https://github.com/4DNucleome/PartSeg/pull/590))\n\n#### \ud83d\udc1b Bug Fixes\n\n- Update build wheels and sdist to have proper version tag ([#583](https://github.com/4DNucleome/PartSeg/pull/583))\n- Fix removing the first measurement entry in the napari Measurement widget ([#584](https://github.com/4DNucleome/PartSeg/pull/584))\n- Fix compatybility bug for conda Pyside2 version ([#595](https://github.com/4DNucleome/PartSeg/pull/595))\n- Error when synchronization is loaded and new iloaded image has different dimensionality than currently loaded. ([#598](https://github.com/4DNucleome/PartSeg/pull/598))\n\n#### \ud83d\ude9c Refactor\n\n- Refactor the create batch plan widgets and add test for it ([#587](https://github.com/4DNucleome/PartSeg/pull/587))\n- Drop napari below 0.4.12 ([#592](https://github.com/4DNucleome/PartSeg/pull/592))\n- Update the order of ROI Mask algorithms to be the same as in older PartSeg versions ([#600](https://github.com/4DNucleome/PartSeg/pull/600))\n\n#### Build\n\n- *(deps)* Bump partsegcore-compiled-backend from 0.13.11 to 0.14.0 in /requirements ([#582](https://github.com/4DNucleome/PartSeg/pull/582))\n- *(deps)* Bump simpleitk from 2.1.1 to 2.1.1.2 in /requirements ([#589](https://github.com/4DNucleome/PartSeg/pull/589))\n- *(deps)* Bump pyinstaller from 4.10 to 5.0 in /requirements ([#586](https://github.com/4DNucleome/PartSeg/pull/586))\n\n### 0.14.0 - 2022-04-14\n\n#### \ud83d\ude80 Features\n\n- Allow to set zoom factor from interface in Search Label napari plugin ([#538](https://github.com/4DNucleome/PartSeg/pull/538))\n- Add controlling of zoom factor of search ROI in main GUI ([#540](https://github.com/4DNucleome/PartSeg/pull/540))\n- Better serialization mechanism allow for declaration data structure migration locally ([#462](https://github.com/4DNucleome/PartSeg/pull/462))\n- Make \\`\\*.obsep\" file possible to load in PartSeg Analysis ([#564](https://github.com/4DNucleome/PartSeg/pull/564))\n- Add option to extract measurement profile or roi extraction profile from batch plan ([#568](https://github.com/4DNucleome/PartSeg/pull/568))\n- Allow import calculation plan from batch result excel file ([#567](https://github.com/4DNucleome/PartSeg/pull/567))\n- Improve error reporting when fail to deserialize data ([#574](https://github.com/4DNucleome/PartSeg/pull/574))\n- Launch PartSeg GUI from napari ([#581](https://github.com/4DNucleome/PartSeg/pull/581))\n\n#### \ud83d\udc1b Bug Fixes\n\n- Fix \"Show selected\" rendering mode in PartSeg ROI Mask ([#565](https://github.com/4DNucleome/PartSeg/pull/565))\n\n#### \ud83d\ude9c Refactor\n\n- Store PartSegImage.Image channels as separated arrays ([#554](https://github.com/4DNucleome/PartSeg/pull/554))\n- Remove deprecated modules. ([#429](https://github.com/4DNucleome/PartSeg/pull/429))\n- Switch serialization backen to `nme` ([#569](https://github.com/4DNucleome/PartSeg/pull/569))\n\n#### \ud83d\udcda Documentation\n\n- Update changelog and add new badges to readme ([#580](https://github.com/4DNucleome/PartSeg/pull/580))\n\n#### \ud83e\uddea Testing\n\n- Add test of creating AboutDialog ([#539](https://github.com/4DNucleome/PartSeg/pull/539))\n- Setup test for python 3.10. Disable class_generator test for this python ([#570](https://github.com/4DNucleome/PartSeg/pull/570))\n\n#### Bugfix\n\n- Add access by operator \\[\\] to pydantic.BaseModel base structures for keep backward compatybility ([#579](https://github.com/4DNucleome/PartSeg/pull/579))\n\n#### Build\n\n- *(deps)* Bump sentry-sdk from 1.5.2 to 1.5.3 in /requirements ([#512](https://github.com/4DNucleome/PartSeg/pull/512))\n- *(deps)* Bump ipython from 8.0.0 to 8.0.1 in /requirements ([#513](https://github.com/4DNucleome/PartSeg/pull/513))\n- *(deps)* Bump pandas from 1.3.5 to 1.4.0 in /requirements ([#514](https://github.com/4DNucleome/PartSeg/pull/514))\n- *(deps)* Bump oiffile from 2021.6.6 to 2022.2.2 in /requirements ([#521](https://github.com/4DNucleome/PartSeg/pull/521))\n- *(deps)* Bump numpy from 1.22.1 to 1.22.2 in /requirements ([#524](https://github.com/4DNucleome/PartSeg/pull/524))\n- *(deps)* Bump tifffile from 2021.11.2 to 2022.2.2 in /requirements ([#523](https://github.com/4DNucleome/PartSeg/pull/523))\n- *(deps)* Bump qtpy from 2.0.0 to 2.0.1 in /requirements ([#522](https://github.com/4DNucleome/PartSeg/pull/522))\n- *(deps)* Bump sentry-sdk from 1.5.3 to 1.5.4 in /requirements ([#515](https://github.com/4DNucleome/PartSeg/pull/515))\n- *(deps)* Bump pyinstaller from 4.8 to 4.10 in /requirements ([#545](https://github.com/4DNucleome/PartSeg/pull/545))\n- *(deps)* Bump pillow from 9.0.0 to 9.0.1 in /requirements ([#549](https://github.com/4DNucleome/PartSeg/pull/549))\n- *(deps)* Bump sphinx from 4.4.0 to 4.5.0 in /requirements ([#561](https://github.com/4DNucleome/PartSeg/pull/561))\n- *(deps)* Bump tifffile from 2022.2.9 to 2022.3.25 in /requirements ([#562](https://github.com/4DNucleome/PartSeg/pull/562))\n- *(deps)* Bump sympy from 1.10 to 1.10.1 in /requirements ([#556](https://github.com/4DNucleome/PartSeg/pull/556))\n- *(deps)* Bump sentry-sdk from 1.5.7 to 1.5.8 in /requirements ([#557](https://github.com/4DNucleome/PartSeg/pull/557))\n\n### 0.13.15\n\n#### Bug Fixes\n\n- Using `translation` instead of `translation_grid` for shifting layers. (#474)\n- Bugs in napari plugins (#478)\n- Missing mask when using roi extraction from napari (#479)\n- Fix segmentation fault on macos machines (#487)\n- Fixes for napari 0.4.13 (#506)\n\n#### Documentation\n\n- Create 0.13.15 release (#511)\n- Add categories and preview page workflow for the napari hub (#489)\n\n#### Features\n\n- Assign properties to mask layer in napari measurement widget (#480)\n\n#### Build\n\n- Bump qtpy from 1.11.3 to 2.0.0 in /requirements (#498)\n- Bump pydantic from 1.8.2 to 1.9.0 in /requirements (#496)\n- Bump sentry-sdk from 1.5.1 to 1.5.2 in /requirements (#497)\n- Bump sphinx from 4.3.1 to 4.3.2 in /requirements (#500)\n- Bump pyinstaller from 4.7 to 4.8 in /requirements (#502)\n- Bump pillow from 8.4.0 to 9.0.0 in /requirements (#501)\n- Bump requests from 2.26.0 to 2.27.1 in /requirements (#495)\n- Bump numpy from 1.21.4 to 1.22.0 in /requirements (#499)\n- Bump numpy from 1.22.0 to 1.22.1 in /requirements (#509)\n- Bump sphinx from 4.3.2 to 4.4.0 in /requirements (#510)\n\n### 0.13.14\n\n#### Bug Fixes\n\n- ROI alternative representation (#471)\n- Change additive to translucent in rendering ROI and Mask (#472)\n\n#### Features\n\n- Add morphological watershed segmentation (#469)\n- Add Bilateral image filter (#470)\n\n### 0.13.13\n\n#### Bug Fixes\n\n- Fix bugs in the generation process of the changelog for release. (#428)\n- Restoring ROI on home button click in compare viewer (#443)\n- Fix Measurement name prefix in bundled PartSeg. (#458)\n- Napari widgets registration in pyinstaller bundle (#465)\n- Hide points button if no points are loaded, hide Mask checkbox if no mask is set (#463)\n- Replace Label data instead of adding/removing layers - fix blending layers (#464)\n\n#### Features\n\n- Add threshold information in layer annotation in the Multiple Otsu ROI extraction method (#430)\n- Add option to select rendering method for ROI (#431)\n- Add callback mechanism to ProfileDict, live update of ROI render parameters (#432)\n- Move the info bar on the bottom of the viewer (#442)\n- Add options to load recent files in multiple files widget (#444)\n- Add ROI annotations as properties to napari labels layer created by ROI Extraction widgets (#445)\n- Add signals to ProfileDict, remove redundant synchronization mechanisms (#449)\n- Allow ignoring updates for 21 days (#453)\n- Save all components if no components selected in mask segmentation (#456)\n- Add modal dialog for search ROI components (#459)\n- Add full measurement support as napari widget (#460)\n- Add search labels as napari widget (#467)\n\n#### Refactor\n\n- Export common code for load/save dialog to one place (#437)\n- Change most of call QFileDialog to more generic code (#440)\n\n#### Testing\n\n- Add test for `PartSeg.common_backend` module (#433)\n\n### 0.13.12\n\n#### Bug Fixes\n\n- Importing the previous version of settings (#406)\n- Cutting without masking data (#407)\n- Save in subdirectory in batch plan (#414)\n- Loading plugins for batch processing (#423)\n\n#### Features\n\n- Add randomization option for correlation calculation (#421)\n- Add Imagej TIFF writer for image. (#405)\n- Mask create widget for napari (#395)\n- In napari roi extraction method show information from roi extraction method (#408)\n- Add `*[0-9].tif` button in batch processing window (#412)\n- Better label representation in 3d view (#418)\n\n#### Refactor\n\n- Use Font Awesome instead of custom symbols (#424)\n\n### 0.13.11\n\n#### Bug Fixes\n\n- Adding mask in Prepare Plan for batch (#383)\n- Set proper completion mode in SearchComboBox (#384)\n- Showing warnings on the error with ROI load (#385)\n\n#### Features\n\n- Add CellFromNucleusFlow \"Cell from nucleus flow\" cell segmentation method (#367)\n- When cutting components in PartSeg ROI mask allow not masking outer data (#379)\n- Theme selection in GUI (#381)\n- Allow return points from ROI extraction algorithm (#382)\n- Add measurement to get ROI annotation by name. (#386)\n- PartSeg ROI extraction algorithms as napari plugins (#387)\n- Add Pearson, Mander's, Intensity, Spearman colocalization measurements (#392)\n- Separate standalone napari settings from PartSeg embedded napari settings (#397)\n\n#### Performance\n\n- Use faster calc bound function (#375)\n\n#### Refactor\n\n- Remove CustomApplication (#389)\n\n### 0.13.10\n\n- change tiff save backend to ome-tiff\n- add `DistanceROIROI` and `ROINeighbourhoodROI` measurements\n\n### 0.13.9\n\n- annotation show bugfix\n\n### 0.13.8\n\n- napari deprecation fixes\n- speedup simple measurement\n- bundle plugins initial support\n\n### 0.13.7\n\n- add measurements widget for napari\n- fix bug in pipeline usage\n\n### 0.13.6\n\n- Hotfix release\n- Prepare for a new napari version\n\n### 0.13.5\n\n- Small fixes for error reporting\n- Fix mask segmentation\n\n### 0.13.4\n\n- Bugfix for outdated profile/pipeline preview\n\n### 0.13.3\n\n- Fix saving roi_info in multiple files and history\n\n### 0.13.2\n\n- Fix showing label in select label tab\n\n### 0.13.1\n\n- Add Haralick measurements\n- Add obsep file support\n\n### 0.13.0\n\n- Add possibility of custom input widgets for algorithms\n- Switch to napari Colormaps instead of custom one\n- Add points visualization\n- Synchronization widget for builtin (View menu) napari viewer\n- Drop Python 3.6\n\n### 0.12.7\n\n- Fixes for napari 0.4.6\n\n### 0.12.6\n\n- Fix prev_mask_get\n- Fix cache mechanism on mask change\n- Update PyInstaller build\n\n### 0.12.5\n\n- Fix bug in pipeline execute\n\n### 0.12.4\n\n- Fix ROI Mask windows related build (signal not properly connected)\n\n### 0.12.3\n\n- Fix ROI Mask\n\n### 0.12.2\n\n- Fix windows bundle\n\n### 0.12.1\n\n- History of last opened files\n- Add ROI annotation and ROI alternatives\n- Minor bugfix\n\n### 0.12.0\n\n- Toggle multiple files widget in View menu\n- Toggle Left panel in ROI Analysis in View Menu\n- Rename Mask Segmentation to ROI Mask\n- Add documentation for interface\n- Add Batch processing tutorial\n- Add information about errors to batch processing output file\n- Load image from the batch prepare window\n- Add search option in part of list and combo boxes\n- Add drag and drop mechanism to load list of files to batch window.\n\n### 0.11.5\n\n- add side view to viewer\n- fix horizontal view for Measurements result table\n\n### 0.11.4\n\n- bump to napari 0.3.8 in bundle\n- fix bug with not presented segmentation loaded from project\n- add frame (1 pix) to image cat from base one based on segmentation\n- pin to Qt version to 5.14\n\n### 0.11.3\n\n- prepare for napari 0.3.7\n- split napari io plugin on multiple part\n- better reporting for numpy array via sentry\n- fix setting color for mask marking\n\n### 0.11.2\n\n- Speedup image set in viewer using async calls\n- Fix bug in long name of sheet with parameters\n\n### 0.11.1\n\n- Add screenshot option in View menu\n- Add Voxels measurements\n\n### 0.11.0\n\n- Make sprawl algorithm name shorter\n- Unify capitalisation of measurement names\n- Add simple measurements to mask segmentation\n- Use napari as viewer\n- Add possibility to preview additional output of algorithms (In View menu)\n- Update names of available Algorithm and Measurement to be more descriptive.\n\n### 0.10.8\n\n- fix synchronisation between viewers in Segmentation Analysis\n- fix batch crash on error during batch run, add information about file on which calculation fails\n- add changelog preview in Help > About\n\n### 0.10.7\n\n- in measurements, on empty list of components mean will return 0\n\n### 0.10.6\n\n- fix border rim preview\n- fix problem with size of image preview\n- zoom with scroll and moving if rectangle zoom is not marked\n\n### 0.10.5\n\n- make PartSeg PEP517 compatible.\n- fix multiple files widget on Windows (path normalisation)\n\n### 0.10.4\n\n- fix slow zoom\n\n### 0.10.3\n\n- deterministic order of elements in batch processing.\n\n### 0.10.2\n\n- bugfixes\n\n### 0.10.1\n\n- bugfixes\n\n### 0.10.0\n\n- Add creating custom label coloring.\n- Change execs interpreter to python 3.7.\n- Add masking operation in Segmentation Mask.\n- Change license to BSD.\n- Allow select root type in batch processing.\n- Add median filter in preview.\n\n### 0.9.7\n\n- fix bug in compare mask\n\n### 0.9.6\n\n- fix bug in loading project with mask\n- upgrade PyInstaller version (bug GHSA-7fcj-pq9j-wh2r)\n\n### 0.9.5\n\n- fix bug in loading project in \"Segmentation analysis\"\n\n### 0.9.4\n\n- read mask segmentation projects\n- choose source type in batch\n- add initial support to OIF and CZI file format\n- extract utils to PartSegCore module\n- add automated tests of example notebook\n- reversed mask\n- load segmentation parameters in mask segmentation\n- allow use sprawl in segmentation tool\n- add radial split of mask for measurement\n- add all measurement results in batch, per component sheet\n\n### 0.9.3\n\n- start automated build documentation\n- change color map backend and allow for user to create custom color map.\n- segmentation compare\n- update test engines\n- support of PySide2\n\n### 0.9.2.3\n\n- refactor code to make easier create plugin for mask segmentation\n- create class base updater for update outdated algorithm description\n- fix save functions\n- fix different bugs\n\n### 0.9.2.2\n\n- extract static data to separated package\n- update marker of fix range and add mark of gauss in channel control\n\n### 0.9.2.1\n\n- add VoteSmooth and add choosing of smooth algorithm\n\n### 0.9.2\n\n- add pypi base check for update\n\n- remove resetting image state when change state in same image\n\n- in stack segmentation add options to picking components from segmentation's\n\n- in mask segmentation add:\n\n - preview of segmentation parameters per component,\n - save segmentation parameters in save file\n - new implementation of batch mode.\n\n### 0.9.1\n\n- Add multiple files widget\n\n- Add Calculating distances between segmented object and mask\n\n- Batch processing plan fixes:\n\n - Fix adding pipelines to plan\n - Redesign mask widget\n\n- modify measurement backend to allow calculate multi channel measurements.\n\n### 0.9\n\nBegin of changelog\n",
"description_content_type": "text/markdown",
"keywords": "bioimaging,GUI",
"home_page": null,
"download_url": null,
"author": null,
"author_email": "Grzegorz Bokota <[email protected]>",
"maintainer": null,
"maintainer_email": null,
"license": "BSD-3-Clause",
"classifier": [
"Development Status :: 3 - Alpha",
"Framework :: napari",
"License :: OSI Approved :: BSD License",
"Operating System :: OS Independent",
"Programming Language :: Python :: 3",
"Programming Language :: Python :: 3 :: Only",
"Programming Language :: Python :: 3.8",
"Programming Language :: Python :: 3.9",
"Programming Language :: Python :: 3.10",
"Programming Language :: Python :: 3.11",
"Programming Language :: Python :: 3.12",
"Programming Language :: Python :: Implementation :: CPython",
"Topic :: Scientific/Engineering :: Bio-Informatics",
"Topic :: Scientific/Engineering :: Image Processing",
"Topic :: Scientific/Engineering :: Visualization"
],
"requires_dist": [
"IPython >=7.7.0",
"PartSegCore-compiled-backend <0.16.0,>=0.13.11",
"PartSegData ==0.10.0",
"QtAwesome !=1.2.0,>=1.0.3",
"QtPy >=1.10.0",
"SimpleITK >=2.0.0",
"appdirs >=1.4.4",
"czifile >=2019.5.22",
"defusedxml >=0.6.0",
"fonticon-fontawesome6 >=6.1.1",
"h5py >=3.3.0",
"imagecodecs >=2020.5.30",
"imageio >=2.5.0",
"ipykernel >=5.2.0",
"local-migrator >=0.1.7",
"magicgui !=0.5.0,>=0.4.0",
"mahotas >=1.4.10",
"napari >=0.4.14",
"nme >=0.1.7",
"oiffile >=2020.1.18",
"openpyxl >=2.5.7",
"packaging >=20.0",
"pandas >=1.1.0",
"psygnal >=0.3.1",
"pydantic <3,>=1.9.1",
"pygments >=2.12.0",
"qtconsole >=4.7.7",
"requests >=2.25.0",
"scipy >=1.4.1",
"sentry-sdk >=2.4.0",
"six >=1.11.0",
"superqt >=0.3.0",
"sympy >=1.1.1",
"tifffile >=2020.9.30",
"traceback-with-variables >=2.0.4",
"vispy >=0.9.4",
"xlrd >=1.1.0",
"xlsxwriter >=2.0.0",
"numpy <2,>=1.18.5 ; python_version < \"3.10\"",
"numpy >=1.18.5 ; python_version >= \"3.10\"",
"PartSeg[accelerate,pyqt5] ; extra == 'all'",
"autodoc-pydantic ; extra == 'docs'",
"sphinx !=3.0.0,!=3.5.0 ; extra == 'docs'",
"sphinx-autodoc-typehints ; extra == 'docs'",
"sphinx-qt-documentation ; extra == 'docs'",
"PartSeg[pyinstaller_base,pyqt5] ; extra == 'pyinstaller'",
"PartSeg[accelerate] ; extra == 'pyinstaller_base'",
"PyInstaller ; extra == 'pyinstaller_base'",
"pydantic ; extra == 'pyinstaller_base'",
"PartSeg[pyqt5] ; extra == 'pyqt'",
"PyQt5 !=5.15.0,>=5.12.3 ; extra == 'pyqt5'",
"napari[pyqt5] ; extra == 'pyqt5'",
"PyQt6 ; extra == 'pyqt6'",
"napari[pyqt6] >=0.5.0 ; (python_version >= \"3.9\") and extra == 'pyqt6'",
"PartSeg[pyside2] ; extra == 'pyside'",
"PySide2 !=5.15.0,>=5.12.3 ; extra == 'pyside2'",
"napari[pyside] ; extra == 'pyside2'",
"PySide6 ; extra == 'pyside6'",
"napari[pyside6_experimental] >=0.5.0 ; (python_version >= \"3.9\") and extra == 'pyside6'",
"coverage ; extra == 'test'",
"lxml[html_clean] ; extra == 'test'",
"pytest >=7.0.0 ; extra == 'test'",
"pytest-qt ; extra == 'test'",
"pytest-timeout ; extra == 'test'",
"scikit-image ; extra == 'test'",
"pytest ; extra == 'testing'",
"pytest-qt ; extra == 'testing'",
"lxml ; extra == 'testing'"
],
"requires_python": ">=3.8",
"requires_external": null,
"project_url": [
"Homepage, https://partseg.github.io/",
"Documentation, https://partseg.readthedocs.io/en/stable/",
"Source Code, https://github.com/4DNucleome/PartSeg",
"User Support, https://github.com/4DNucleome/PartSeg/issues",
"Bug Tracker, https://github.com/4DNucleome/PartSeg/issues"
],
"provides_extra": [
"accelerate",
"all",
"docs",
"pyinstaller",
"pyinstaller_base",
"pyqt",
"pyqt5",
"pyqt6",
"pyside",
"pyside2",
"pyside6",
"test",
"testing"
],
"provides_dist": null,
"obsoletes_dist": null
},
"npe1_shim": false
}