-
Notifications
You must be signed in to change notification settings - Fork 0
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
How to get past the gotchas without getting gotten again #2
Comments
|
Whenever moving back to using scipiper / remake, I almost always forget to use |
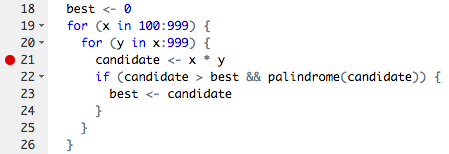
How to inspect parts of the pipeline and variables within functionsIf you've written your own functions or scripts before, you may have run into the red breakpoint dot 🔴 on the left side oyour script window: Breakpoints allow you to run a function (or script) up until the line of the breakpoint, and then the evaluation pauses. You are able to inspect all variables available at that point in the evaluation, and even step carefully forward one line at a time. It is out of scope of this exercise to go through exactly how to use debuggers, but they are powerful and helpful tools. It would be a good idea to read up on them if you haven't run into breakpoints yet. In You have a working, albeit brittle, pipeline in your course repository. You can try it out with So, if you wanted to look at what Then, running ⌨️ comment on where you think you might find I'll sit patiently until you comment |
|
|
Visualizing and understanding the status of dependencies in a pipelineSeeing the structure of a pipeline as a visual is powerful. Viewing connections between targets and the direction data is flowing in can help you better understand the role of pipelines in data science work. Once you are more familiar with pipelines, using the same visuals can help you diagnose problems. Below is a remakefile that is very similar to the one you have in your code repository (the targets:
all:
depends: 3_visualize/out/figure_1.png
site_data:
command: download_nwis_data()
1_fetch/out/site_info.csv:
command: nwis_site_info(fileout = '1_fetch/out/site_info.csv', site_data)
1_fetch/out/nwis_01427207_data.csv:
command: download_nwis_site_data('1_fetch/out/nwis_01427207_data.csv')
1_fetch/out/nwis_01435000_data.csv:
command: download_nwis_site_data('1_fetch/out/nwis_01435000_data.csv')
site_data_clean:
command: process_data(site_data)
site_data_annotated:
command: annotate_data(site_data_clean, site_filename = '1_fetch/out/site_info.csv')
site_data_styled:
command: style_data(site_data_annotated)
3_visualize/out/figure_1.png:
command: plot_nwis_timeseries(fileout = '3_visualize/out/figure_1.png',
site_data_styled, width = 12, height = 7, units = I('in'))Two file targets ( remake::diagram()The remake::diagram()If you run the same command, you'll see something similar but the two new files won't be included. Seeing this diagram helps develop a greater understanding of some of the earlier concepts from intro-to-pipelines. Here, you can clearly see the connection between The diagram also shows how the inputs of one function create connections to the output of that function. By modifying the recipe for the all target, it is possible to create a dependency link to one of the .csv files, which would then result in that file being included in a build as it becomes necessary in order to complete targets:
all:
depends: ["3_visualize/out/figure_1.png", "1_fetch/out/nwis_01427207_data.csv"]And after calling With this update, the build of ⌨️ comment on what you learned from exploring I'll sit patiently until you comment |
|
|
|
|
What are cyclical dependencies and how to avoid them?using As a reminder, the direction of the arrows capture the dependency flow, and Also note that there are no backward looking arrows. What if This potentially infinite loop is confusing to think about and is also something that dependency managers can't support. We won't say much more about this issue here, but note that in the early days of building pipelines if you run into a cyclical dependency error, this is what's going on. ⌨️ Add a comment when you are ready to move on. I'll sit patiently until you comment |
|
Once we talked about putting in an infinite while loop in Brian Weidel's R environment that would say "Hi Brian", whenever he opened R. We didn't implement but had some good laughs |
Creating side-effect targets or undocumented inputsMoving into a pipeline-way-of-thinking can reveal some suprising habits you created when working under a different paradigm. Moving the work of scripts into functions is one thing that helps compartmentalize thinking and organize data and code relationships, but smart pipelines require even more special attention to how functions are designed. side-effect targetsIt is tempting to build functions that do several things; perhaps a plotting function also writes a table, or a data munging function returns a data.frame, but also writes a log file. You may have noticed that there is no easy way to specify two ouputs from a single function in a pipeline recipe. We can have multiple files as inputs into a function/command, but only one output/target. If a function writes a file that is not explicitly connected in the recipe (i.e., it is a "side-effect" output), the file is untracked by the dependency manager, and treated like an implicit file target (i.e., a target which has no Maybe the above doesn't sound like a real issue, since the side-effect target would be updated every time the other explicit target it is paired with is rebuilt. But this becomes a scary problem (and our first real gotcha!) if the explicit target is not connected to the critical path of the final sets of targets you want to build, but the implicit side-effect target is. What this means is that even if the explicit target is out of date, it will not be rebuilt because building this target is unnecessary to completing the final targets (remember "skip the work you don't need" ↪️). The dependency manager doesn't know that there is a hidden rule for updating the side-effect target and that this update is necessary for assuring the final targets are up-to-date and correct. 🔀 Side-effect targets can be used effectively, but doing so requires a good understanding of implications for tracking them and advanced strategies on how to specify rules and dependencies in a way that carries them along. ☑️ undocumented inputsAdditionally, it is tempting to code a filepath within a function which has information that needs to be accessed in order to run. This seems harmless, since functions are tracked by the dependency manager and any changes to those will trigger rebuilds, right? Not quite. If a file like As a rule, unless you are purposefully trying to hide changes in a file from the dependency manager, do not read non-argument files in the body of a function. 🔚 ⌨️ Add a comment when you are ready to move on I'll sit patiently until you comment |
|
👍 |
How (not to) depend on a directory for changesYou might have a project where there is a directory 📁 with a collection of files. To simplify the example, assume all of the files are In a data pipeline, we'd want assurance that any time the number of files changes, we'd rebuild the resulting data.frame. Likewise, if at any point the contents of any one of the files changes, we'd also want to re-build the data.frame. This hypothetical example could be coded as sources:
- combine_files.R
targets:
all:
depends: figure_1.png
plot_data:
command: combine_into_df('1_fetch/in/file1.csv','1_fetch/in/file2.csv','1_fetch/in/file3.csv')
figure_1.png:
command: my_plot(plot_data)☝️ This coding would work, as it tells the dependency manager which files to track for changes, and where the files in the directory are. But this solution is less than ideal, both because it doesn't scale well to many files, and because it doesn't adapt to new files coming into the Alternatively, what about adding the directory as an input to the recipe, like this (you'd also need to modify your sources:
- combine_files.R
targets:
all:
depends: figure_1.png
plot_data:
command: combine_into_df(work_dir = '1_fetch/in')
figure_1.png:
command: my_plot(plot_data)After running An explaination of that warning message - In order to determine if file contents have changed, remake uses
A third strategy might be to create a target that lists the contents of the directory, and then uses that list as an input: sources:
- combine_files.R
targets:
all:
depends: figure_1.png
work_files:
command: dir('1_fetch/in')
plot_data:
command: combine_into_df(work_files)
figure_1.png:
command: my_plot(plot_data)☝️ This approach is close, but has a few flaws: 1) because '1_fetch/in' can't be tracked as a real target, changes within that directory aren't tracked (same issue as the previous example), and 2) if the contents of the files change, but the number and names of the files don't change, the Instead, we would avocate for a modified approach that combines the work of Now, if the But unfortunately, because
⌨️ Add a comment when you are ready to move on. I'll sit patiently until you comment |
|
👍 |
What to do when you want to specify a non-build-object input to a function?Wow, we've gotten this far and haven't written a function that accepts anything other than an object target or a file target. I feel so constrained! In reality, R functions have all kinds of other arguments, from logicals (TRUE/FALSE), to characters that specify which configurations to use. The example in your working pipeline creates a figure, called But adding those to the remake file like so 3_visualize/out/figure_1.png:
command: plot_nwis_timeseries(target_name, site_data_styled, width = 12, height = 7, units = 'in')causes an immediate error Since We know 3_visualize/out/figure_1.png:
command: plot_nwis_timeseries(target_name, site_data_styled, width = 12, height = 7, units = I('in'))works! 🌟 Going back to our previous example where we wanted to build a data.frame of file hashes from the '1_fetch/in' directory, sources:
- combine_files.R
targets:
all:
depends: figure_1.png
work_files:
command: hash_dir_files(directory = I('1_fetch/in'), dummy = I('2020-05-18'))By adding this ⌨️ Add a comment when you are ready to move on. I'll sit patiently until you comment |
|
👍 |
The
|
|
I'm pretty sure |
When you are done poking around, check out the next issue. |
Thanks for the reminder, Jake. @jread-usgs , what do you think about adding in some info about target_name argument ordering to the section above? I think the remake issue I created (richfitz/remake#173) isn't quite complete - if I remember right, you actually can use target_name out of order as long as you don't name any of the arguments - but that's a pretty restrictive condition, and the general guideline of placing target_name first seems like the best way to keep everyone out of trouble & frustration. |
|
huh, interesting. I never ran into that issue (or forgot about it) and have used I added to the remake issue, but will copy here. Note that this is possible when your function supports named arguments. In the example above the args to file.create are ..., showWarnings = TRUE. I agree that it would be nice that target_name could be found when unnamed, it does work when you can tie it to an argument name: target_default: i.txt
targets:
i.txt:
command: file.create(target_name, showWarnings=FALSE)
i2.txt:
command: file.copy(from = 'i.txt', to = target_name)works with |
|
Ah, that's good to know. |
|
Back with this
Was that this same issue/error "object 'target_name' not found" or were you getting weirder errors because you weren't naming the arguments and were assuming an order than the function itself wasn't using? |
|
To be honest I don't remember the exact error as it was probably ~2 years ago - it was probably supplying unnamed arguments w/ |




In this section, we're going to go one by one through a series of tips that will help you avoid common pitfalls (or gotchas!) in pipelines. These tips will help you in the next sections and in future work. A quick list of what's to come:
which_dirty()andwhy_dirty()to further interrogate the status of pipeline targetsI()helpertarget_namespecial variable. Simplifyingtargetcommandrelationships and reducing duplication⌨️ add a comment to this issue and the bot will respond with the next topic
I'll sit patiently until you comment
The text was updated successfully, but these errors were encountered: