diff --git a/models/vista2d/configs/metadata.json b/models/vista2d/configs/metadata.json
index 9a870f64..fcec60da 100644
--- a/models/vista2d/configs/metadata.json
+++ b/models/vista2d/configs/metadata.json
@@ -1,7 +1,8 @@
{
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20240725.json",
- "version": "0.2.6",
+ "version": "0.2.7",
"changelog": {
+ "0.2.7": "enhance readme",
"0.2.6": "update tensorrt benchmark results",
"0.2.5": "add tensorrt benchmark results",
"0.2.4": "enable tensorrt inference",
diff --git a/models/vista2d/docs/README.md b/models/vista2d/docs/README.md
index 38b1a26f..4fd639a5 100644
--- a/models/vista2d/docs/README.md
+++ b/models/vista2d/docs/README.md
@@ -6,7 +6,6 @@ A pretrained model was trained on collection of 15K public microscopy images. Th
-
### Model highlights
- Robust deep learning algorithm based on transformers
@@ -15,10 +14,9 @@ A pretrained model was trained on collection of 15K public microscopy images. Th
- Multiple modalities of imaging data collectively supported
- Multi-GPU and multinode training support
-
### Generalization performance
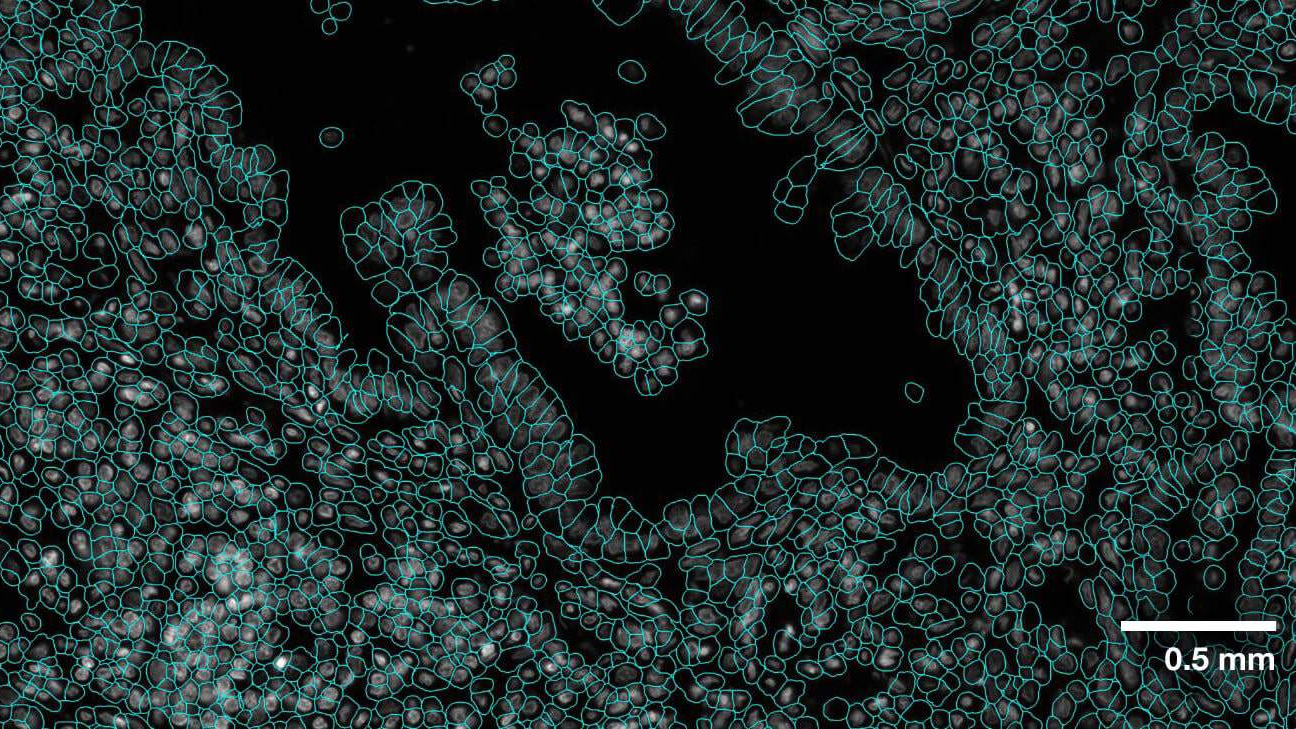
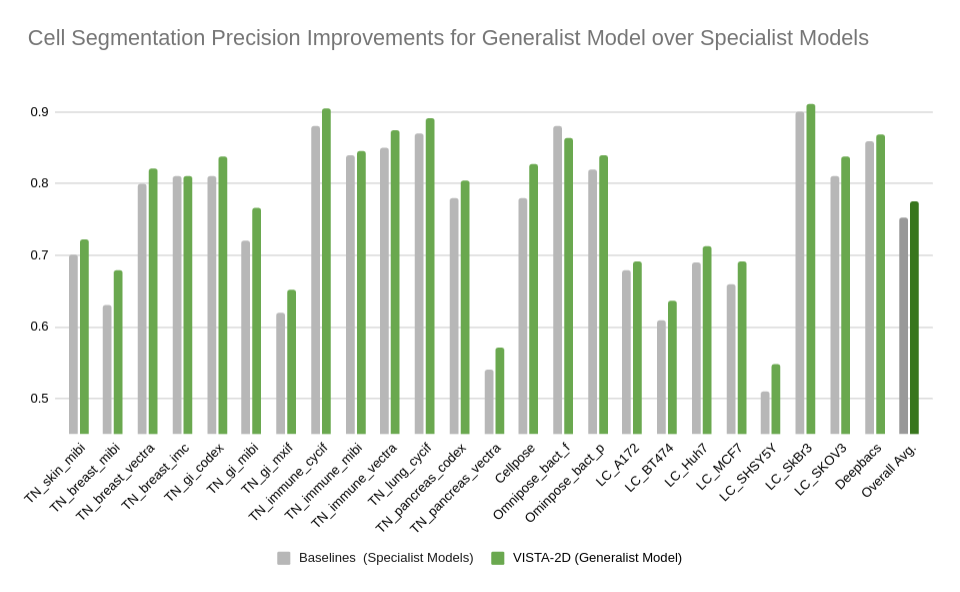
-Evaluation was performed for the VISTA2D model with multiple public datasets, such as TissueNet, LIVECell, Omnipose, DeepBacs, Cellpose, and [more](./docs/data_license.txt). A total of ~15K annotated cell images were collected to train the generalist VISTA2D model. This ensured broad coverage of many different types of cells, which were acquired by various imaging acquisition types. The benchmark results of the experiment were performed on held-out test sets for each public dataset that were already defined by the dataset contributors. Average precision at an IoU threshold of 0.5 was used for evaluating performance. The benchmark results are reported in comparison with the best numbers found in the literature, in addition to a specialist VISTA2D model trained only on a particular dataset or a subset of data.
+Evaluation was performed for the VISTA2D model with multiple public datasets, such as TissueNet, LIVECell, Omnipose, DeepBacs, Cellpose, and more. For more details about dataset licenses, please refer to `/docs/data_license.txt`. A total of ~15K annotated cell images were collected to train the generalist VISTA2D model. This ensured broad coverage of many different types of cells, which were acquired by various imaging acquisition types. The benchmark results of the experiment were performed on held-out test sets for each public dataset that were already defined by the dataset contributors. Average precision at an IoU threshold of 0.5 was used for evaluating performance. The benchmark results are reported in comparison with the best numbers found in the literature, in addition to a specialist VISTA2D model trained only on a particular dataset or a subset of data.
@@ -49,10 +47,31 @@ This result is benchmarked under:
### Prepare Data Lists and Datasets
-The default dataset for training, validation, and inference is the [Cellpose](https://www.cellpose.org/) dataset. Please follow the [tutorial](../download_preprocessor/) to prepare the dataset before executing any commands below.
+The default dataset for training, validation, and inference is the [Cellpose](https://www.cellpose.org/) dataset. Please follow the `download_preprocessor/` to prepare the dataset before executing any commands below.
Additionally, all data lists are available in the `datalists.zip` file located in the root directory of the bundle. Extract the contents of the `.zip` file to access the data lists.
+### Dependencies
+Please refer to `required_packages_version` in `configs/metadata.json` to install all necessary dependencies before executing.
+
+Important Note: if your environment already contains OpenCV, installing `cellpose` may lead to conflicts and produce errors such as:
+
+```
+AttributeError: partially initialized module 'cv2' has no attribute 'dnn' (most likely due to a circular import)
+```
+
+when executing. To resolve this issue, please uninstall OpenCV and then re-install `cellpose` with a command like:
+
+```Bash
+pip uninstall -y opencv && rm /usr/local/lib/python3.x/dist-packages/cv2
+```
+
+Alternatively, you can use the following command to install `cellpose` without its dependencies:
+
+```
+pip install --no-deps cellpose
+```
+
### Execute training
```bash
python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml
@@ -192,7 +211,7 @@ Ask and answer questions on [MONAI VISTA's GitHub discussions tab](https://githu
## License
-The codebase is under Apache 2.0 Licence. The model weight is released under CC-BY-NC-SA-4.0. For various public data licenses please see [data_license.txt](data_license.txt).
+The codebase is under Apache 2.0 Licence. The model weight is released under CC-BY-NC-SA-4.0. For various public data licenses please see `data_license.txt`.
## Acknowledgement
- [segment-anything](https://github.com/facebookresearch/segment-anything)
diff --git a/models/vista2d/download_preprocessor/readme.md b/models/vista2d/download_preprocessor/readme.md
index e07cc8be..4c60ed16 100644
--- a/models/vista2d/download_preprocessor/readme.md
+++ b/models/vista2d/download_preprocessor/readme.md
@@ -54,7 +54,7 @@ The `datasets` directory needs to be selected as highlighted in the screenshot,
a user created directory named `omnipose_dataset`.
### The remaining datasets will be downloaded by a python script.
-To run the script use the following example command `python all_file_downloader.py --download_path provide_the_same_root_data_path`
+To run the script use the following example command `python all_file_downloader.py --dir provide_the_same_root_data_path`
After completion of downloading of all datasets, below is how the data root directory should look:
@@ -63,11 +63,11 @@ After completion of downloading of all datasets, below is how the data root dire
### Process the downloaded data
To execute VISTA-2D training pipeline, some datasets require label conversion. Please use the `root_data_path` as the input to the script, example command to execute the script is given below:
-`python generate_json.py --data_root provide_the_same_root_data_path`
+`python generate_json.py --dir provide_the_same_root_data_path`
### Generation of Json data lists (Optional)
If one desires to generate JSON files from scratch, `generate_json.py` script performs both processing and creation of JSON files.
To execute VISTA-2D training pipeline, some datasets require label conversion and then a json file list which the VISTA-2D training uses a format.
Creating the json lists from the raw dataset sources, please use the `root_data_path` as the input to the script, example command to execute the script is given below:
-`python generate_json.py --data_root provide_the_same_root_data_path`
+`python generate_json.py --dir provide_the_same_root_data_path`
diff --git a/models/vista3d/configs/inference.json b/models/vista3d/configs/inference.json
index 46bd6ff2..92f2c0dd 100644
--- a/models/vista3d/configs/inference.json
+++ b/models/vista3d/configs/inference.json
@@ -124,7 +124,7 @@
"inferer": {
"_target_": "scripts.inferer.Vista3dInferer",
"roi_size": "@patch_size",
- "overlap": 0.5,
+ "overlap": 0.3,
"sw_batch_size": "@sw_batch_size",
"use_point_window": "@use_point_window"
},
diff --git a/models/vista3d/configs/metadata.json b/models/vista3d/configs/metadata.json
index 1255e7ca..be73909a 100644
--- a/models/vista3d/configs/metadata.json
+++ b/models/vista3d/configs/metadata.json
@@ -1,7 +1,8 @@
{
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20240725.json",
- "version": "0.4.7",
+ "version": "0.4.8",
"changelog": {
+ "0.4.8": "use 0.3 overlap for inference",
"0.4.7": "update tensorrt benchmark results",
"0.4.6": "add tensorrt benchmark result and remove the metric part",
"0.4.5": "remove wrong path",