Changelog
CARLISLE 2.6.0¶
Bug fixes¶
- Bug fixes for DESeq (#127, @epehrsson)
- Removes single-sample group check for DESeq.
- Increases memory for DESeq.
- Ensures control replicate number is an integer.
- Fixes FDR cutoff misassigned to log2FC cutoff.
- Fixes
no_dedupvariable names in library normalization scripts. - Fig bug that added nonexistent directories to the singularity bind paths. (#135, @kelly-sovacool)
- Containerize rules that require R (
deseq,go_enrichment, andspikein_assessment) to fix installation issues with common R library path. (#129, @kelly-sovacool)- The
Rlib_dirandRpkg_configconfig options have been removed as they are no longer needed.
- The
New features¶
- New visualizations: (#132, @epehrsson)
- New rules
cov_correlation,homer_enrich,combine_homer,count_peaks - Add peak caller to MACS2 peak xls filename
- New parameters in the config file to make certain rules optional: (#133, @kelly-sovacool)
- GO enrichment is controlled by
run_go_enrichment(default:false) - ROSE is controlled by
run_rose(default:false) - New
--singcacheargument to provide a singularity cache dir location. The singularity cache dir is automatically set inside/data/$USER/or$WORKDIR/if--singcacheis not provided. (#143, @kelly-sovacool)
Misc¶
- The singularity version is no longer specified, per request of the biowulf admins. (#139, @kelly-sovacool)
- Minor documentation updates. (#146, @kelly-sovacool)
CARLISLE v2.5.0¶
- Refactors R packages to a common source location (#118, @slsevilla)
- Adds a --force flag to allow for re-initialization of a workdir (#97, @slsevilla)
- Fixes error with testrun in DESEQ2 (#113, @slsevilla)
- Decreases the number of samples being run with testrun, essentially running tinytest as default and removing tinytest as an option (#115, @slsevilla)
- Reads version from VERSION file instead of github repo link (#96, #112, @slsevilla)
- Added a CHANGELOG (#116, @slsevilla)
- Fix: RNA report bug, caused by hard-coding of PC1-3, when only PC1-2 were generated (#104, @slsevilla)
- Minor documentation improvements. (#100, @kelly-sovacool)
- Fix: allow printing the version or help message even if singularity is not in the path. (#110, @kelly-sovacool)
CARLISLE v2.4.1¶
- Add GitHub Action to add issues/PRs to personal project boards by @kelly-sovacool in #95
- Create install script by @kelly-sovacool in #93
- feat: use summits bed for homer input; save temporary files; fix deseq2 bug by @slsevilla in #108
- docs: adding citation and DOI to pipeline by @slsevilla in #107
- Test a dryrun with GitHub Actions by @kelly-sovacool in #94
CARLISLE v2.4.0¶
- Feature- Merged Yue's fork, adding DEEPTOOLS by @slsevilla in #85
- Feature- Added tracking features from SPOOK by @slsevilla in #88
- Feature - Dev test run completed by @slsevilla in #89
- Bug - Fixed bugs related to Biowulf transition
CARLISLE v2.1.0¶
- enhancement
- update gopeaks resources
- change SEACR to run "norm" without spikein controls, "non" with spikein controls
- update docs for changes; provide extra troubleshooting guidance
- fix GoEnrich bug for failed plots
CARLISLE v2.0.1¶
- fix error when contrasts set to "N"
- adjust goenrich resources to be more efficient
CARLISLE 2.0.0¶
- Add a MAPQ filter to samtools (rule align)
- Add GoPeaks MultiQC module
- Allow for library normalization to occur during first pass
- Add --broad-cutoff to MACS2 broad peak calling for MACS2
- Create a spike in QC report
- Reorganize file structure to help with qthreshold folder
- Update variable names of all peak caller
- Merge rules with input/output/wildcard congruency
- Convert the "spiked" variable to "norm_method
- Add name of control used to MACS2 peaks
- Running extra control:sample comparisons that are not needed
- improved resource allocation
- test data originally included 1475 jobs, this version includes 1087 jobs (reduction of 25%) despite including additional features
- moved ~12% of all jobs to local deployment (within SLURM submission)
CARLISLE 1.2.0¶
- merge increases to resources; update workflow img, contributions
CARLISLE 1.1.1¶
- patch for gz bigbed bug
CARLISLE 1.1.0¶
- add broad-cutoff to macs2 broad peaks param settings
- add non.stringent and non.relaxed to annotation options
- merge DESEQ and DESEQ2 rules together
- identify some files as temp
CARLISLE 1.0.1¶
- contains patch for DESEQ error with non hs1 reference samples
Contributing to CARLISLE¶
Proposing changes with issues¶
If you want to make a change, it's a good idea to first open an issue and make sure someone from the team agrees that it’s needed.
If you've decided to work on an issue, assign yourself to the issue so others will know you're working on it.
Pull request process¶
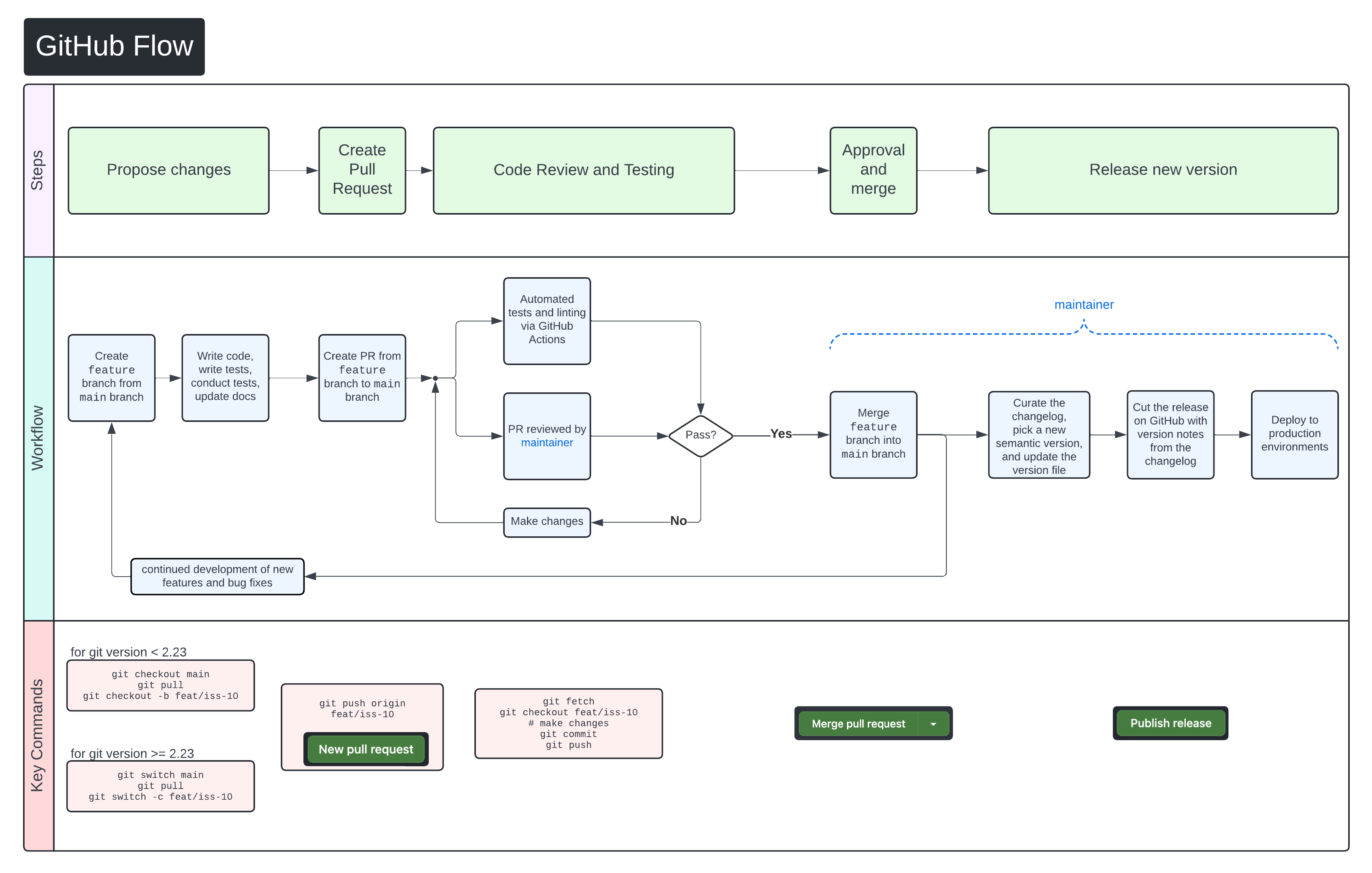
We use GitHub Flow as our collaboration process. Follow the steps below for detailed instructions on contributing changes to CARLISLE.
Clone the repo¶
If you are a member of CCBR, you can clone this repository to your computer or development environment. Otherwise, you will first need to fork the repo and clone your fork. You only need to do this step once.
git clone https://github.com/CCBR/CARLISLE
+Cloning into 'CARLISLE'...
remote: Enumerating objects: 1136, done.
remote: Counting objects: 100% (463/463), done.
remote: Compressing objects: 100% (357/357), done.
remote: Total 1136 (delta 149), reused 332 (delta 103), pack-reused 673
Receiving objects: 100% (1136/1136), 11.01 MiB | 9.76 MiB/s, done.
Resolving deltas: 100% (530/530), done.
cd CARLISLE
+If this is your first time cloning the repo, you may need to install dependencies¶
-
Install snakemake and singularity or docker if needed (biowulf already has these available as modules).
-
Install the python dependencies with pip
pip install .
+If you're developing on biowulf, you can use our shared conda environment which already has these dependencies installed
. "/data/CCBR_Pipeliner/db/PipeDB/Conda/etc/profile.d/conda.sh"
+conda activate py311
+- Install
pre-commitif you don't already have it. Then from the repo's root directory, run
pre-commit install
+This will install the repo's pre-commit hooks. You'll only need to do this step the first time you clone the repo.
Create a branch¶
Create a Git branch for your pull request (PR). Give the branch a descriptive name for the changes you will make, such as iss-10 if it is for a specific issue.
# create a new branch and switch to it
+git branch iss-10
+git switch iss-10
+Switched to a new branch 'iss-10'
Make your changes¶
Edit the code, write and run tests, and update the documentation as needed.
test¶
Changes to the python package code will also need unit tests to demonstrate that the changes work as intended. We write unit tests with pytest and store them in the tests/ subdirectory. Run the tests with python -m pytest.
If you change the workflow, please run the workflow with the test profile and make sure your new feature or bug fix works as intended.
document¶
If you have added a new feature or changed the API of an existing feature, you will likely need to update the documentation in docs/.
Commit and push your changes¶
If you're not sure how often you should commit or what your commits should consist of, we recommend following the "atomic commits" principle where each commit contains one new feature, fix, or task. Learn more about atomic commits here: https://www.freshconsulting.com/insights/blog/atomic-commits/
First, add the files that you changed to the staging area:
git add path/to/changed/files/
+Then make the commit. Your commit message should follow the Conventional Commits specification. Briefly, each commit should start with one of the approved types such as feat, fix, docs, etc. followed by a description of the commit. Take a look at the Conventional Commits specification for more detailed information about how to write commit messages.
git commit -m 'feat: create function for awesome feature'
+pre-commit will enforce that your commit message and the code changes are styled correctly and will attempt to make corrections if needed.
Check for added large files..............................................Passed
Fix End of Files.........................................................Passed
Trim Trailing Whitespace.................................................Failed
- hook id: trailing-whitespace
- exit code: 1
- files were modified by this hook
>
Fixing path/to/changed/files/file.txt
>
codespell................................................................Passed
style-files..........................................(no files to check)Skipped
readme-rmd-rendered..................................(no files to check)Skipped
use-tidy-description.................................(no files to check)Skipped
In the example above, one of the hooks modified a file in the proposed commit, so the pre-commit check failed. You can run git diff to see the changes that pre-commit made and git status to see which files were modified. To proceed with the commit, re-add the modified file(s) and re-run the commit command:
git add path/to/changed/files/file.txt
+git commit -m 'feat: create function for awesome feature'
+This time, all the hooks either passed or were skipped (e.g. hooks that only run on R code will not run if no R files were committed). When the pre-commit check is successful, the usual commit success message will appear after the pre-commit messages showing that the commit was created.
Check for added large files..............................................Passed
Fix End of Files.........................................................Passed
Trim Trailing Whitespace.................................................Passed
codespell................................................................Passed
style-files..........................................(no files to check)Skipped
readme-rmd-rendered..................................(no files to check)Skipped
use-tidy-description.................................(no files to check)Skipped
Conventional Commit......................................................Passed
> [iss-10 9ff256e] feat: create function for awesome feature
1 file changed, 22 insertions(+), 3 deletions(-)
Finally, push your changes to GitHub:
git push
+If this is the first time you are pushing this branch, you may have to explicitly set the upstream branch:
git push --set-upstream origin iss-10
+Enumerating objects: 7, done.
Counting objects: 100% (7/7), done.
Delta compression using up to 10 threads
Compressing objects: 100% (4/4), done.
Writing objects: 100% (4/4), 648 bytes | 648.00 KiB/s, done.
Total 4 (delta 3), reused 0 (delta 0), pack-reused 0
remote: Resolving deltas: 100% (3/3), completed with 3 local objects.
remote:
remote: Create a pull request for 'iss-10' on GitHub by visiting:
remote: https://github.com/CCBR/CARLISLE/pull/new/iss-10
remote:
To https://github.com/CCBR/CARLISLE
>
> [new branch] iss-10 -> iss-10
branch 'iss-10' set up to track 'origin/iss-10'.
We recommend pushing your commits often so they will be backed up on GitHub. You can view the files in your branch on GitHub at https://github.com/CCBR/CARLISLE/tree/<your-branch-name> (replace <your-branch-name> with the actual name of your branch).
Create the PR¶
Once your branch is ready, create a PR on GitHub: https://github.com/CCBR/CARLISLE/pull/new/
Select the branch you just pushed:
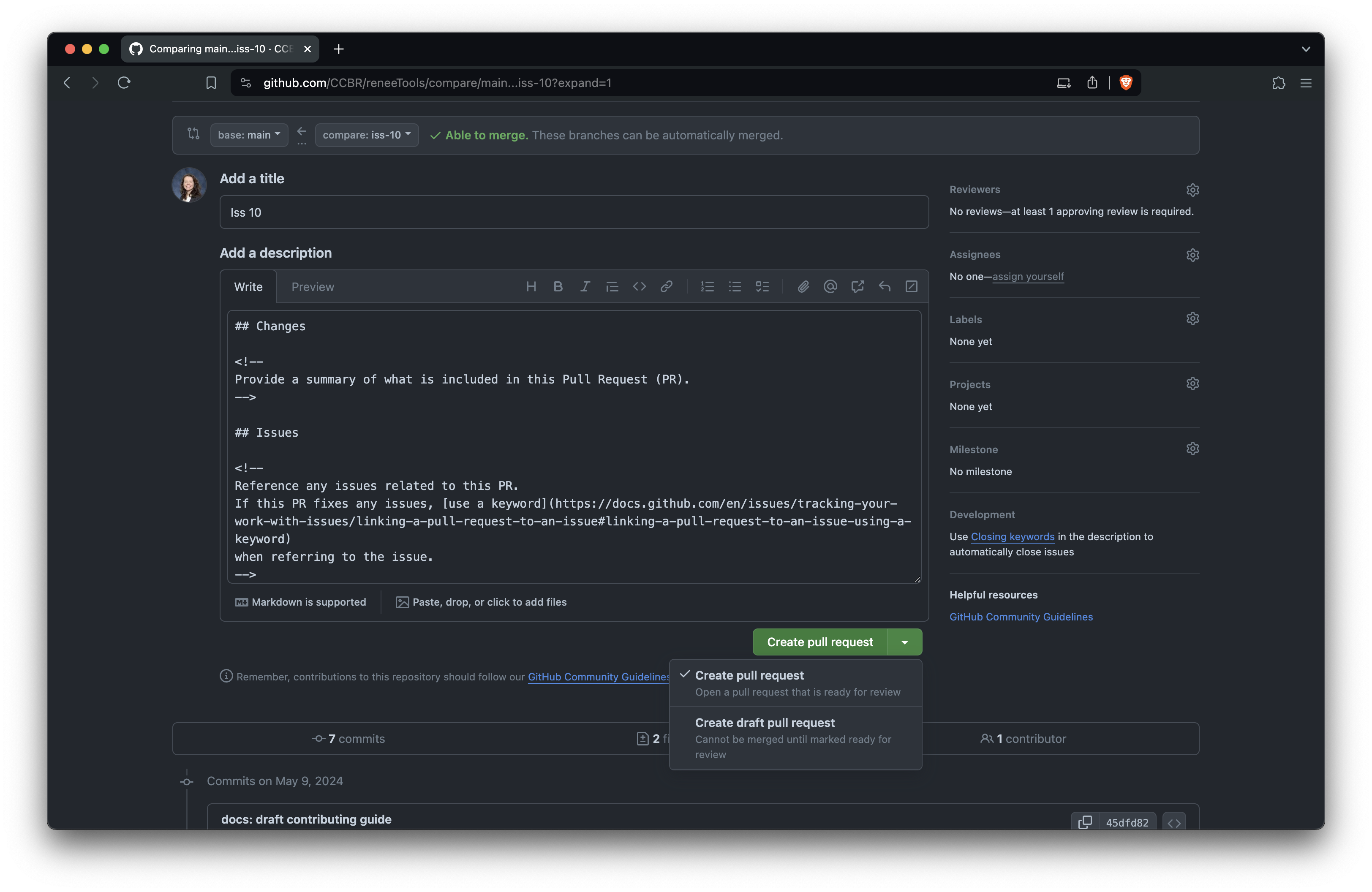
Edit the PR title and description. The title should briefly describe the change. Follow the comments in the template to fill out the body of the PR, and you can delete the comments (everything between <!-- and -->) as you go. Be sure to fill out the checklist, checking off items as you complete them or striking through any irrelevant items. When you're ready, click 'Create pull request' to open it.
Optionally, you can mark the PR as a draft if you're not yet ready for it to be reviewed, then change it later when you're ready.
Wait for a maintainer to review your PR¶
We will do our best to follow the tidyverse code review principles: https://code-review.tidyverse.org/. The reviewer may suggest that you make changes before accepting your PR in order to improve the code quality or style. If that's the case, continue to make changes in your branch and push them to GitHub, and they will appear in the PR.
Once the PR is approved, the maintainer will merge it and the issue(s) the PR links will close automatically. Congratulations and thank you for your contribution!
After your PR has been merged¶
After your PR has been merged, update your local clone of the repo by switching to the main branch and pulling the latest changes:
git checkout main
+git pull
+It's a good idea to run git pull before creating a new branch so it will start from the most recent commits in main.
Helpful links for more information¶
- GitHub Flow
- semantic versioning guidelines
- changelog guidelines
- tidyverse code review principles
- reproducible examples
- nf-core extensions for VS Code
CARLISLE¶
Cut And Run anaLysIS pipeLinE
This snakemake pipeline is built to run on Biowulf.
For comments/suggestions/advice please contact CCBR_Pipeliner@mail.nih.gov.
For detailed documentation on running the pipeline view the documentation website.
Workflow¶
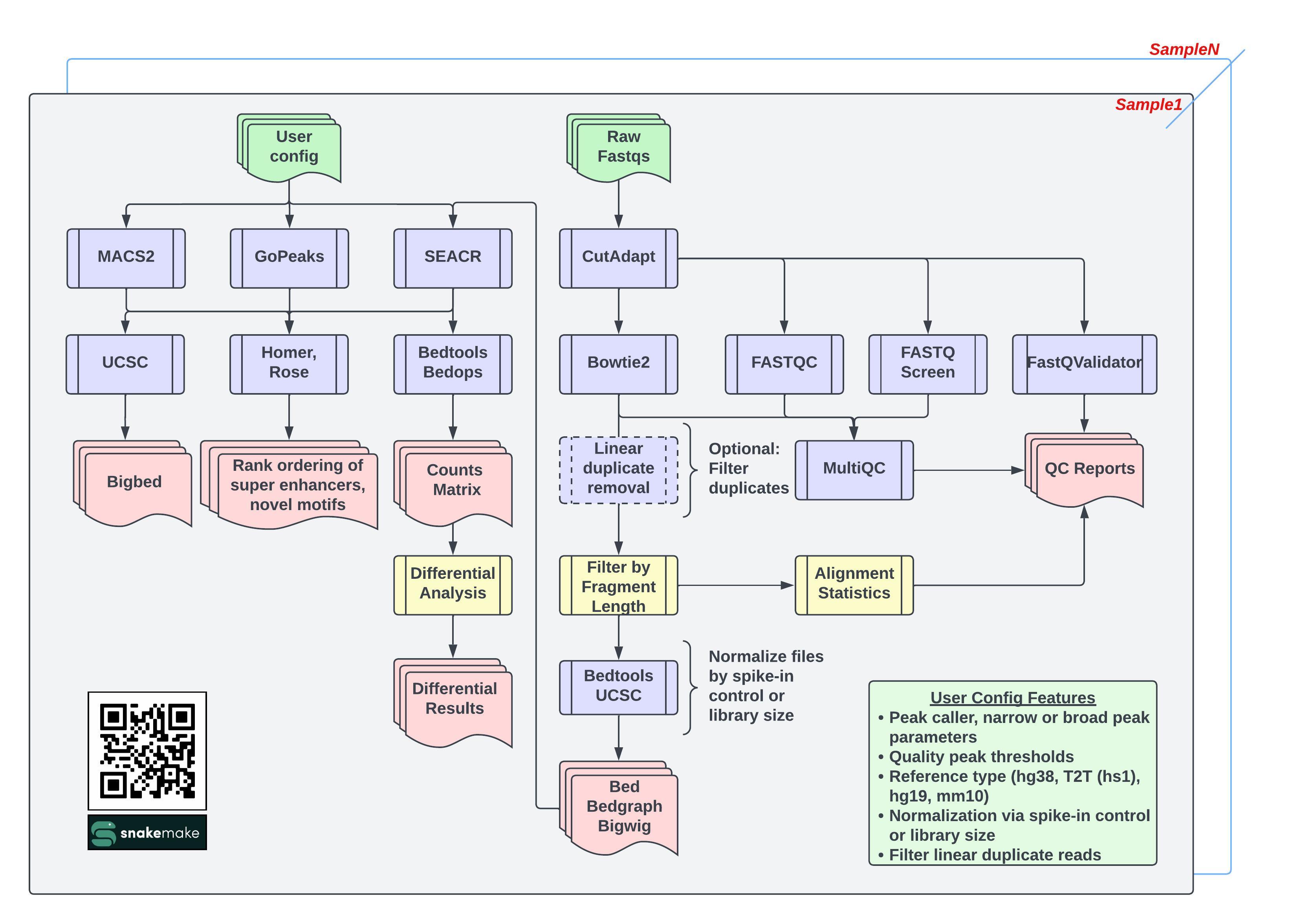
The CARLISLE pipeline was developed in support of NIH Dr Vassiliki Saloura's Laboratory and Dr Javed Khan's Laboratory. It has been developed and tested solely on NIH HPC Biowulf.
Cut And Run anaLysIS pipeLinE
This snakemake pipeline is built to run on Biowulf.
For comments/suggestions/advice please contact CCBR_Pipeliner@mail.nih.gov.
For detailed documentation on running the pipeline view the documentation website.
"},{"location":"#workflow","title":"Workflow","text":"The CARLISLE pipeline was developed in support of NIH Dr Vassiliki Saloura's Laboratory and Dr Javed Khan's Laboratory. It has been developed and tested solely on NIH HPC Biowulf.
"},{"location":"changelog/","title":"Changelog","text":""},{"location":"changelog/#carlisle-260","title":"CARLISLE 2.6.0","text":""},{"location":"changelog/#bug-fixes","title":"Bug fixes","text":"- Bug fixes for DESeq (#127, @epehrsson)
- Removes single-sample group check for DESeq.
- Increases memory for DESeq.
- Ensures control replicate number is an integer.
- Fixes FDR cutoff misassigned to log2FC cutoff.
- Fixes
no_dedupvariable names in library normalization scripts. - Fig bug that added nonexistent directories to the singularity bind paths. (#135, @kelly-sovacool)
- Containerize rules that require R (
deseq,go_enrichment, andspikein_assessment) to fix installation issues with common R library path. (#129, @kelly-sovacool)- The
Rlib_dirandRpkg_configconfig options have been removed as they are no longer needed.
- The
- New visualizations: (#132, @epehrsson)
- New rules
cov_correlation,homer_enrich,combine_homer,count_peaks - Add peak caller to MACS2 peak xls filename
- New parameters in the config file to make certain rules optional: (#133, @kelly-sovacool)
- GO enrichment is controlled by
run_go_enrichment(default:false) - ROSE is controlled by
run_rose(default:false) - New
--singcacheargument to provide a singularity cache dir location. The singularity cache dir is automatically set inside/data/$USER/or$WORKDIR/if--singcacheis not provided. (#143, @kelly-sovacool)
- The singularity version is no longer specified, per request of the biowulf admins. (#139, @kelly-sovacool)
- Minor documentation updates. (#146, @kelly-sovacool)
- Refactors R packages to a common source location (#118, @slsevilla)
- Adds a --force flag to allow for re-initialization of a workdir (#97, @slsevilla)
- Fixes error with testrun in DESEQ2 (#113, @slsevilla)
- Decreases the number of samples being run with testrun, essentially running tinytest as default and removing tinytest as an option (#115, @slsevilla)
- Reads version from VERSION file instead of github repo link (#96, #112, @slsevilla)
- Added a CHANGELOG (#116, @slsevilla)
- Fix: RNA report bug, caused by hard-coding of PC1-3, when only PC1-2 were generated (#104, @slsevilla)
- Minor documentation improvements. (#100, @kelly-sovacool)
- Fix: allow printing the version or help message even if singularity is not in the path. (#110, @kelly-sovacool)
- Add GitHub Action to add issues/PRs to personal project boards by @kelly-sovacool in #95
- Create install script by @kelly-sovacool in #93
- feat: use summits bed for homer input; save temporary files; fix deseq2 bug by @slsevilla in #108
- docs: adding citation and DOI to pipeline by @slsevilla in #107
- Test a dryrun with GitHub Actions by @kelly-sovacool in #94
- Feature- Merged Yue's fork, adding DEEPTOOLS by @slsevilla in #85
- Feature- Added tracking features from SPOOK by @slsevilla in #88
- Feature - Dev test run completed by @slsevilla in #89
- Bug - Fixed bugs related to Biowulf transition
- enhancement
- update gopeaks resources
- change SEACR to run \"norm\" without spikein controls, \"non\" with spikein controls
- update docs for changes; provide extra troubleshooting guidance
- fix GoEnrich bug for failed plots
- fix error when contrasts set to \"N\"
- adjust goenrich resources to be more efficient
- Add a MAPQ filter to samtools (rule align)
- Add GoPeaks MultiQC module
- Allow for library normalization to occur during first pass
- Add --broad-cutoff to MACS2 broad peak calling for MACS2
- Create a spike in QC report
- Reorganize file structure to help with qthreshold folder
- Update variable names of all peak caller
- Merge rules with input/output/wildcard congruency
- Convert the \"spiked\" variable to \"norm_method
- Add name of control used to MACS2 peaks
- Running extra control:sample comparisons that are not needed
- improved resource allocation
- test data originally included 1475 jobs, this version includes 1087 jobs (reduction of 25%) despite including additional features
- moved ~12% of all jobs to local deployment (within SLURM submission)
- merge increases to resources; update workflow img, contributions
- patch for gz bigbed bug
- add broad-cutoff to macs2 broad peaks param settings
- add non.stringent and non.relaxed to annotation options
- merge DESEQ and DESEQ2 rules together
- identify some files as temp
- contains patch for DESEQ error with non hs1 reference samples
If you want to make a change, it's a good idea to first open an issue and make sure someone from the team agrees that it\u2019s needed.
If you've decided to work on an issue, assign yourself to the issue so others will know you're working on it.
"},{"location":"contributing/#pull-request-process","title":"Pull request process","text":"We use GitHub Flow as our collaboration process. Follow the steps below for detailed instructions on contributing changes to CARLISLE.
"},{"location":"contributing/#clone-the-repo","title":"Clone the repo","text":"If you are a member of CCBR, you can clone this repository to your computer or development environment. Otherwise, you will first need to fork the repo and clone your fork. You only need to do this step once.
git clone https://github.com/CCBR/CARLISLE\nCloning into 'CARLISLE'... remote: Enumerating objects: 1136, done. remote: Counting objects: 100% (463/463), done. remote: Compressing objects: 100% (357/357), done. remote: Total 1136 (delta 149), reused 332 (delta 103), pack-reused 673 Receiving objects: 100% (1136/1136), 11.01 MiB | 9.76 MiB/s, done. Resolving deltas: 100% (530/530), done.
cd CARLISLE\n-
Install snakemake and singularity or docker if needed (biowulf already has these available as modules).
-
Install the python dependencies with pip
pip install .\nIf you're developing on biowulf, you can use our shared conda environment which already has these dependencies installed
. \"/data/CCBR_Pipeliner/db/PipeDB/Conda/etc/profile.d/conda.sh\"\nconda activate py311\n- Install
pre-commitif you don't already have it. Then from the repo's root directory, run
pre-commit install\nThis will install the repo's pre-commit hooks. You'll only need to do this step the first time you clone the repo.
"},{"location":"contributing/#create-a-branch","title":"Create a branch","text":"Create a Git branch for your pull request (PR). Give the branch a descriptive name for the changes you will make, such as iss-10 if it is for a specific issue.
# create a new branch and switch to it\ngit branch iss-10\ngit switch iss-10\nSwitched to a new branch 'iss-10'
"},{"location":"contributing/#make-your-changes","title":"Make your changes","text":"Edit the code, write and run tests, and update the documentation as needed.
"},{"location":"contributing/#test","title":"test","text":"Changes to the python package code will also need unit tests to demonstrate that the changes work as intended. We write unit tests with pytest and store them in the tests/ subdirectory. Run the tests with python -m pytest.
If you change the workflow, please run the workflow with the test profile and make sure your new feature or bug fix works as intended.
"},{"location":"contributing/#document","title":"document","text":"If you have added a new feature or changed the API of an existing feature, you will likely need to update the documentation in docs/.
If you're not sure how often you should commit or what your commits should consist of, we recommend following the \"atomic commits\" principle where each commit contains one new feature, fix, or task. Learn more about atomic commits here: https://www.freshconsulting.com/insights/blog/atomic-commits/
First, add the files that you changed to the staging area:
git add path/to/changed/files/\nThen make the commit. Your commit message should follow the Conventional Commits specification. Briefly, each commit should start with one of the approved types such as feat, fix, docs, etc. followed by a description of the commit. Take a look at the Conventional Commits specification for more detailed information about how to write commit messages.
git commit -m 'feat: create function for awesome feature'\npre-commit will enforce that your commit message and the code changes are styled correctly and will attempt to make corrections if needed.
Check for added large files..............................................Passed Fix End of Files.........................................................Passed Trim Trailing Whitespace.................................................Failed
- hook id: trailing-whitespace
- exit code: 1
- files were modified by this hook > Fixing path/to/changed/files/file.txt > codespell................................................................Passed style-files..........................................(no files to check)Skipped readme-rmd-rendered..................................(no files to check)Skipped use-tidy-description.................................(no files to check)Skipped
In the example above, one of the hooks modified a file in the proposed commit, so the pre-commit check failed. You can run git diff to see the changes that pre-commit made and git status to see which files were modified. To proceed with the commit, re-add the modified file(s) and re-run the commit command:
git add path/to/changed/files/file.txt\ngit commit -m 'feat: create function for awesome feature'\nThis time, all the hooks either passed or were skipped (e.g. hooks that only run on R code will not run if no R files were committed). When the pre-commit check is successful, the usual commit success message will appear after the pre-commit messages showing that the commit was created.
Check for added large files..............................................Passed Fix End of Files.........................................................Passed Trim Trailing Whitespace.................................................Passed codespell................................................................Passed style-files..........................................(no files to check)Skipped readme-rmd-rendered..................................(no files to check)Skipped use-tidy-description.................................(no files to check)Skipped Conventional Commit......................................................Passed > [iss-10 9ff256e] feat: create function for awesome feature 1 file changed, 22 insertions(+), 3 deletions(-)
Finally, push your changes to GitHub:
git push\nIf this is the first time you are pushing this branch, you may have to explicitly set the upstream branch:
git push --set-upstream origin iss-10\nEnumerating objects: 7, done. Counting objects: 100% (7/7), done. Delta compression using up to 10 threads Compressing objects: 100% (4/4), done. Writing objects: 100% (4/4), 648 bytes | 648.00 KiB/s, done. Total 4 (delta 3), reused 0 (delta 0), pack-reused 0 remote: Resolving deltas: 100% (3/3), completed with 3 local objects. remote: remote: Create a pull request for 'iss-10' on GitHub by visiting: remote: https://github.com/CCBR/CARLISLE/pull/new/iss-10 remote: To https://github.com/CCBR/CARLISLE > > [new branch] iss-10 -> iss-10 branch 'iss-10' set up to track 'origin/iss-10'.
We recommend pushing your commits often so they will be backed up on GitHub. You can view the files in your branch on GitHub at https://github.com/CCBR/CARLISLE/tree/<your-branch-name> (replace <your-branch-name> with the actual name of your branch).
Once your branch is ready, create a PR on GitHub: https://github.com/CCBR/CARLISLE/pull/new/
Select the branch you just pushed:
Edit the PR title and description. The title should briefly describe the change. Follow the comments in the template to fill out the body of the PR, and you can delete the comments (everything between <!-- and -->) as you go. Be sure to fill out the checklist, checking off items as you complete them or striking through any irrelevant items. When you're ready, click 'Create pull request' to open it.
Optionally, you can mark the PR as a draft if you're not yet ready for it to be reviewed, then change it later when you're ready.
"},{"location":"contributing/#wait-for-a-maintainer-to-review-your-pr","title":"Wait for a maintainer to review your PR","text":"We will do our best to follow the tidyverse code review principles: https://code-review.tidyverse.org/. The reviewer may suggest that you make changes before accepting your PR in order to improve the code quality or style. If that's the case, continue to make changes in your branch and push them to GitHub, and they will appear in the PR.
Once the PR is approved, the maintainer will merge it and the issue(s) the PR links will close automatically. Congratulations and thank you for your contribution!
"},{"location":"contributing/#after-your-pr-has-been-merged","title":"After your PR has been merged","text":"After your PR has been merged, update your local clone of the repo by switching to the main branch and pulling the latest changes:
git checkout main\ngit pull\nIt's a good idea to run git pull before creating a new branch so it will start from the most recent commits in main.
- GitHub Flow
- semantic versioning guidelines
- changelog guidelines
- tidyverse code review principles
- reproducible examples
- nf-core extensions for VS Code
The following members contributed to the development of the CARLISLE pipeline:
- Samantha Sevilla
- Vishal Koparde
- Hsien-chao Chou
- Sohyoung Kim
- Yue Hu
- Vassiliki Saloura
VK, SS, SK, HC contributed to the generating the source code and all members contributed to the main concepts and analysis.
"},{"location":"user-guide/getting-started/","title":"Overview","text":"The CARLISLE github repository is stored locally, and will be used for project deployment. Multiple projects can be deployed from this one point simultaneously, without concern.
"},{"location":"user-guide/getting-started/#1-getting-started","title":"1. Getting Started","text":""},{"location":"user-guide/getting-started/#11-introduction","title":"1.1 Introduction","text":"The CARLISLE Pipelie beings with raw FASTQ files and performs trimming followed by alignment using BOWTIE2. Data is then normalized through either the use of an user-species species (IE E.Coli) spike-in control or through the determined library size. Peaks are then called using MACS2, SEACR, and GoPEAKS with various options selected by the user. Peaks are then annotated, and summarized into reports. If designated, differential analysis is performed using DESEQ2. QC reports are also generated with each project using FASTQC and MULTIQC. Annotations are added using HOMER and ROSE. GSEA Enrichment analysis predictions are added using CHIPENRICH.
The following are sub-commands used within CARLISLE:
- initialize: initalize the pipeline
- dryrun: predict the binding of peptides to any MHC molecule
- cluster: execute the pipeline on the Biowulf HPC
- local: execute a local, interactive, session
- git: execute GitHub actions
- unlock: unlock directory
- DAG: create DAG report
- report: create SNAKEMAKE report
- runtest: copies test manifests and files to WORKDIR
CARLISLE has several dependencies listed below. These dependencies can be installed by a sysadmin. All dependencies will be automatically loaded if running from Biowulf.
- bedtools: \"bedtools/2.30.0\"
- bedops: \"bedops/2.4.40\"
- bowtie2: \"bowtie/2-2.4.2\"
- cutadapt: \"cutadapt/1.18\"
- fastqc: \"fastqc/0.11.9\"
- fastq_screen: \"fastq_screen/0.15.2\"
- fastq_val: \"/data/CCBR_Pipeliner/iCLIP/bin/fastQValidator\"
- fastxtoolkit: \"fastxtoolkit/0.0.14\"
- gopeaks: \"github clone https://github.com/maxsonBraunLab/gopeaks\"
- macs2: \"macs/2.2.7.1\"
- multiqc: \"multiqc/1.9\"
- perl: \"perl/5.34.0\"
- picard: \"picard/2.26.9\"
- python37: \"python/3.7\"
- R: \"R/4.2.2\"
- rose: \"ROSE/1.3.1\"
- samtools: \"samtools/1.15\"
- seacr: \"seacr/1.4-beta.2\"
- ucsc: \"ucsc/407\"
CARLISLE has been exclusively tested on Biowulf HPC. Login to the cluster's head node and move into the pipeline location.
# ssh into cluster's head node\nssh -Y $USER@biowulf.nih.gov\nAn interactive session should be started before performing any of the pipeline sub-commands, even if the pipeline is to be executed on the cluster.
# Grab an interactive node\nsinteractive --time=12:00:00 --mem=8gb --cpus-per-task=4 --pty bash\nThe following directories are created under the WORKDIR/results directory:
- alignment_stats: this directory include information on the alignment of each sample
- bam: this directory includes BAM files, statistics on samples, statistics on spike-in controls for each sample
- bedgraph: this directory includes BEDGRAPH files and statistic summaries for each sample
- bigwig: this directory includes the bigwig files for each sample
- peaks: this directory contains a sub-directory that relates to the quality threshold used.
- quality threshold
- contrasts: this directory includes the contrasts for each line listed in the contrast manifest
- peak_caller: this directory includes all peak calls from each peak_caller (SEACR, MACS2, GOPEAKS) for each sample
- annotation
- go_enrichment: this directory includes gene set enrichment pathway predictions when
run_go_enrichmentis set totruein the config file. - homer: this directory includes the annotation output from HOMER
- rose: this directory includes the annotation output from ROSE when
run_roseis set totruein the config file.
- go_enrichment: this directory includes gene set enrichment pathway predictions when
- qc: this directory includes MULTIQC reports and spike-in control reports (when applicable)
\u251c\u2500\u2500 alignment_stats\n\u251c\u2500\u2500 bam\n\u251c\u2500\u2500 bedgraph\n\u251c\u2500\u2500 bigwig\n\u251c\u2500\u2500 fragments\n\u251c\u2500\u2500 peaks\n\u2502\u00a0\u00a0 \u251c\u2500\u2500 0.05\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 contrasts\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 contrast_id1.dedup_status\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 contrast_id2.dedup_status\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 gopeaks\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 annotation\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 go_enrichment\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 contrast_id1.dedup_status.go_enrichment_tables\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 contrast_id2.dedup_status.go_enrichment_html_report\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homer\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.gopeaks_broad.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.gopeaks_narrow.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.gopeaks_broad.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.gopeaks_narrow.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 rose\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.gopeaks_broad.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.gopeaks_narrow.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.dedup.gopeaks_broad.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.dedup.gopeaks_narrow.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 peak_output\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 macs2\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 annotation\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 go_enrichment\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 contrast_id1.dedup_status.go_enrichment_tables\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 contrast_id2.dedup_status.go_enrichment_html_report\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homer\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.macs2_narrow.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.macs2_broad.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.macs2_narrow.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.macs2_broad.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 rose\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.macs2_broad.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.macs2_narrow.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.macs2_broad.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.macs2_narrow.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 peak_output\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 seacr\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 annotation\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 go_enrichment\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 contrast_id1.dedup_status.go_enrichment_tables\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 contrast_id2.dedup_status.go_enrichment_html_report\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homer\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.seacr_non_relaxed.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.seacr_non_stringent.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.seacr_norm_relaxed.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.seacr_norm_stringent.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.seacr_non_relaxed.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.seacr_non_stringent.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.seacr_norm_relaxed.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.seacr_norm_stringent.motifs\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 homerResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 knownResults\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 rose\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.seacr_non_relaxed.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.seacr_non_stringent.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.seacr_norm_relaxed.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id1_vs_control_id.dedup_status.seacr_norm_stringent.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.seacr_non_relaxed.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.seacr_non_stringent.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.seacr_norm_relaxed.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 replicate_id2_vs_control_id.dedup_status.seacr_norm_stringent.12500\n\u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 peak_output\n\u2514\u2500\u2500 qc\n \u251c\u2500\u2500 fastqc_raw\n \u2514\u2500\u2500 fqscreen_raw\nThe pipeline is controlled through editing configuration and manifest files. Defaults are found in the /WORKDIR/config and /WORKDIR/manifest directories, after initialization.
"},{"location":"user-guide/preparing-files/#21-configs","title":"2.1 Configs","text":"The configuration files control parameters and software of the pipeline. These files are listed below:
- config/config.yaml
- resources/cluster.yaml
- resources/tools.yaml
The cluster configuration file dictates the resouces to be used during submission to Biowulf HPC. There are two differnt ways to control these parameters - first, to control the default settings, and second, to create or edit individual rules. These parameters should be edited with caution, after significant testing.
"},{"location":"user-guide/preparing-files/#212-tools-config","title":"2.1.2 Tools Config","text":"The tools configuration file dictates the version of each software or program that is being used in the pipeline.
"},{"location":"user-guide/preparing-files/#213-config-yaml","title":"2.1.3 Config YAML","text":"There are several groups of parameters that are editable for the user to control the various aspects of the pipeline. These are :
- Folders and Paths
- These parameters will include the input and ouput files of the pipeline, as well as list all manifest names.
- User parameters
- These parameters will control the pipeline features. These include thresholds and whether to perform processes.
- References
- These parameters will control the location of index files, spike-in references, adaptors and species calling information.
The pipeline allows for the use of a species specific spike-in control, or the use of normalization via library size. The parameter spikein_genome should be set to the species term used in spikein_reference.
For example for ecoli spike-in:
run_contrasts: true\nnorm_method: \"spikein\"\nspikein_genome: \"ecoli\"\nspikein_reference:\n ecoli:\n fa: \"PIPELINE_HOME/resources/spikein/Ecoli_GCF_000005845.2_ASM584v2_genomic.fna\"\nFor example for drosophila spike-in:
run_contrasts: true\nnorm_method: \"spikein\"\nspikein_genome: \"drosophila\"\nspikein_reference:\n drosophila:\n fa: \"/fdb/igenomes/Drosophila_melanogaster/UCSC/dm6/Sequence/WholeGenomeFasta/genome.fa\"\nIf it's determined that the amount of spike-in is not sufficient for the run, a library normaliaztion can be performed.
- Complete a CARLISLE run with spike-in set to \"Y\". This will allow for the complete assessment of the spike-in.
- Run inital QC analysis on the output data
- Add the alignment_stats dir to the configuration file.
- Re-run the CARLISLE pipeline
Users can select duplicated peaks (dedup) or non-deduplicated peaks (no_dedup) through the user parameter.
dupstatus: \"dedup, no_dedup\"\nThree peak callers are available for deployment within the pipeline, with different settings deployed for each caller.
- MACS2 is available with two peak calling options: narrowPeak or broadPeak. NOTE: DESeq step generally fails for broadPeak; generally has too many calls.
peaktype: \"macs2_narrow, macs2_broad,\"\n- SEACR is available with four peak calling options: stringent or relaxed parameters, to be paired with \"norm\" for samples without a spike-in control and \"non\" for samples with a spikein control
peaktype: \"seacr_stringent, seacr_relaxed\"\n- GOPEAKS is available with two peak calling options: narrowpeaks or broadpeaks
peaktype: \"gopeaks_narrow, gopeaks_broad\"\nA complete list of the available peak calling parameters and the recommended list of parameters is provided below:
Peak Caller Narrow Broad Normalized, Stringent Normalized, Relaxed Non-Normalized, Stringent Non-Normalized, Relaxed Macs2 AVAIL AVAIL NA NA NA NA SEACR NA NA AVAIL w/o SPIKEIN AVAIL w/o SPIKEIN AVAIL w/ SPIKEIN AVAIL w/ SPIKEIN GoPeaks AVAIL AVAIL NA NA NA NA# Recommended list\n### peaktype: \"macs2_narrow, macs2_broad, gopeaks_narrow, gopeaks_broad\"\n\n# Available list\n### peaktype: \"macs2_narrow, macs2_broad, seacr_norm_stringent, seacr_norm_relaxed, seacr_non_stringent, seacr_non_relaxed, gopeaks_narrow, gopeaks_broad\"\nMACS2 can be run with or without the control. adding a control will increase peak specificity Selecting \"Y\" for the macs2_control will run the paired control sample provided in the sample manifest
Thresholds for quality can be controled through the quality_tresholds parameter. This must be a list of comma separated values. minimum of numeric value required.
- default MACS2 qvalue is 0.05 https://manpages.ubuntu.com/manpages/xenial/man1/macs2_callpeak.1.html
- default GOPEAKS pvalue is 0.05 https://github.com/maxsonBraunLab/gopeaks/blob/main/README.md
- default SEACR FDR threshold 1 https://github.com/FredHutch/SEACR/blob/master/README.md
#default values\nquality_thresholds: \"0.1, 0.05, 0.01\"\nAdditional reference files may be added to the pipeline, if other species were to be used.
The absolute file paths which must be included are:
- fa: \"/path/to/species.fa\"
- blacklist: \"/path/to/blacklistbed/species.bed\"
The following information must be included:
- regions: \"list of regions to be included; IE chr1 chr2 chr3\"
- macs2_g: \"macs2 genome shorthand; IE mm IE hs\"
There are two manifests, one which required for all pipeliens and one that is only required if running a differential analysis. These files describe information on the samples and desired contrasts. The paths of these files are defined in the snakemake_config.yaml file. These files are:
- samplemanifest
- contrasts
This manifest will include information to sample level information. It includes the following column headers:
- sampleName: the sample name WITHOUT replicate number (IE \"SAMPLE\")
- replicateNumber: the sample replicate number (IE \"1\")
- isControl: whether the sample should be identified as a control (IE \"Y\")
- controlName: the name of the control to use for this sample (IE \"CONTROL\")
- controlReplicateNumber: the replicate number of the control to use for this sample (IE \"1\")
- path_to_R1: the full path to R1 fastq file (IE \"/path/to/sample1.R1.fastq\")
- path_to_R2: the full path to R1 fastq file (IE \"/path/to/sample2.R2.fastq\")
An example sampleManifest file is shown below:
sampleName replicateNumber isControl controlName controlReplicateNumber path_to_R1 path_to_R2 53_H3K4me3 1 N HN6_IgG_rabbit_negative_control 1 PIPELINE_HOME/.test/53_H3K4me3_1.R1.fastq.gz PIPELINE_HOME/.test/53_H3K4me3_1.R2.fastq.gz 53_H3K4me3 2 N HN6_IgG_rabbit_negative_control 1 PIPELINE_HOME/.test/53_H3K4me3_2.R1.fastq.gz PIPELINE_HOME/.test/53_H3K4me3_2.R2.fastq.gz HN6_H3K4me3 1 N HN6_IgG_rabbit_negative_control 1 PIPELINE_HOME/.test/HN6_H3K4me3_1.R1.fastq.gz PIPELINE_HOME/.test/HN6_H3K4me3_1.R2.fastq.gz HN6_H3K4me3 2 N HN6_IgG_rabbit_negative_control 1 PIPELINE_HOME/.test/HN6_H3K4me3_2.R1.fastq.gz PIPELINE_HOME/.test/HN6_H3K4me3_2.R2.fastq.gz HN6_IgG_rabbit_negative_control 1 Y - - PIPELINE_HOME/.test/HN6_IgG_rabbit_negative_control_1.R1.fastq.gz PIPELINE_HOME/.test/HN6_IgG_rabbit_negative_control_1.R2.fastq.gz"},{"location":"user-guide/preparing-files/#222-contrast-manifest-optional","title":"2.2.2 Contrast Manifest (OPTIONAL)","text":"This manifest will include sample information to performed differential comparisons.
An example contrast file:
condition1 condition2 MOC1_siSmyd3_2m_25_HCHO MOC1_siNC_2m_25_HCHONote: you must have more than one sample per condition in order to perform differential analysis with DESeq2
"},{"location":"user-guide/run/","title":"3. Running the Pipeline","text":""},{"location":"user-guide/run/#31-pipeline-overview","title":"3.1 Pipeline Overview","text":"The Snakemake workflow has a multiple options
"},{"location":"user-guide/run/#required-arguments","title":"Required arguments","text":"Usage: bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle -m/--runmode=<RUNMODE> -w/--workdir=<WORKDIR>\n\n1. RUNMODE: [Type: String] Valid options:\n *) init : initialize workdir\n *) run : run with slurm\n *) reset : DELETE workdir dir and re-init it\n *) dryrun : dry run snakemake to generate DAG\n *) unlock : unlock workdir if locked by snakemake\n *) runlocal : run without submitting to sbatch\n *) runtest: run on cluster with included test dataset\n2. WORKDIR: [Type: String]: Absolute or relative path to the output folder with write permissions.\n--help|-h : print this help. --version|-v : print the version of carlisle. --force|-f : use the force flag for snakemake to force all rules to run. --singcache|-c : singularity cache directory. Default is /data/${USER}/.singularity if available, or falls back to ${WORKDIR}/.singularity. Use this flag to specify a different singularity cache directory.
The following explains each of the command options:
- Preparation Commands
- init (REQUIRED): This must be performed before any Snakemake run (dry, local, cluster) can be performed. This will copy the necessary config, manifest and Snakefiles needed to run the pipeline to the provided output directory.
- the -f/--force flag can be used in order to re-initialize a workdir that has already been created
- dryrun (OPTIONAL): This is an optional step, to be performed before any Snakemake run (local, cluster). This will check for errors within the pipeline, and ensure that you have read/write access to the files needed to run the full pipeline.
- Processing Commands
- local: This will run the pipeline on a local node. NOTE: This should only be performed on an interactive node.
- run: This will submit a master job to the cluster, and subsequent sub-jobs as needed to complete the workflow. An email will be sent when the pipeline begins, if there are any errors, and when it completes.
- Other Commands (All optional)
- unlock: This will unlock the pipeline if an error caused it to stop in the middle of a run.
- runtest: This will run a test of the pipeline with test data
To run any of these commands, follow the the syntax:
bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=COMMAND --workdir=/path/to/output/dir\nA typical command workflow, running on the cluser, is as follows:
bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=init --workdir=/path/to/output/dir\n\nbash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=dryrun --workdir=/path/to/output/dir\n\nbash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=run --workdir=/path/to/output/dir\nWelcome to the CARLISLE Pipeline Tutorial!
"},{"location":"user-guide/test-info/#51-getting-started","title":"5.1 Getting Started","text":"Review the information on the Getting Started for a complete overview the pipeline. The tutorial below will use test data available on NIH Biowulf HPC only. All example code will assume you are running v1.0 of the pipeline, using test data available on GitHub.
A. Change working directory to the CARLISLE repository
B. Initialize Pipeline
bash ./path/to/dir/carlisle --runmode=init --workdir=/path/to/output/dir\nTest data is included in the .test directory as well as the config directory.
A Run the test command to prepare the data, perform a dry-run and submit to the cluster
bash ./path/to/dir/carlisle --runmode=runtest --workdir=/path/to/output/dir\n- An expected output for the
runtestis as follows:
Job stats:\njob count min threads max threads\n----------------------------- ------- ------------- -------------\nDESeq 24 1 1\nalign 9 56 56\nalignstats 9 2 2\nall 1 1 1\nbam2bg 9 4 4\ncreate_contrast_data_files 24 1 1\ncreate_contrast_peakcaller_files 12 1 1\ncreate_reference 1 32 32\ncreate_replicate_sample_table 1 1 1\ndiffbb 24 1 1\nfilter 18 2 2\nfindMotif 96 6 6\ngather_alignstats 1 1 1\ngo_enrichment 12 1 1\ngopeaks_broad 16 2 2\ngopeaks_narrow 16 2 2\nmacs2_broad 16 2 2\nmacs2_narrow 16 2 2\nmake_counts_matrix 24 1 1\nmultiqc 2 1 1\nqc_fastqc 9 1 1\nrose 96 2 2\nseacr_relaxed 16 2 2\nseacr_stringent 16 2 2\nspikein_assessment 1 1 1\ntrim 9 56 56\ntotal 478 1 56\nReview the expected outputs on the Output page. If there are errors, review and performing stesp described on the Troubleshooting page as needed.
"},{"location":"user-guide/troubleshooting/","title":"Troubleshooting","text":"Recommended steps to troubleshoot the pipeline.
"},{"location":"user-guide/troubleshooting/#11-email","title":"1.1 Email","text":"Check your email for an email regarding pipeline failure. You will receive an email from slurm@biowulf.nih.gov with the subject: Slurm Job_id=[#] Name=CARLISLE Failed, Run time [time], FAILED, ExitCode 1
"},{"location":"user-guide/troubleshooting/#12-review-the-log-files","title":"1.2 Review the log files","text":"Review the logs in two ways:
- Review the master slurm file: This file will be found in the
/path/to/results/dir/and titledslurm-[jobid].out. Reviewing this file will tell you what rule errored, and for any local SLURM jobs, provide error details - Review the individual rule log files: After reviewing the master slurm-file, review the specific rules that failed within the
/path/to/results/dir/logs/. Each rule will include a.errand.outfile, with the following formatting:{rulename}.{masterjobID}.{individualruleID}.{wildcards from the rule}.{out or err}
After addressing the issue, unlock the output directory, perform another dry-run and check the status of the pipeline, then resubmit to the cluster.
#unlock dir\nbash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=unlock --workdir=/path/to/output/dir\n\n#perform dry-run\nbash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=dryrun --workdir=/path/to/output/dir\n\n#submit to cluster\nbash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=run --workdir=/path/to/output/dir\nIf after troubleshooting, the error cannot be resolved, or if a bug is found, please create an issue and send and email to Samantha Chill.
"}]} \ No newline at end of file diff --git a/2.6/sitemap.xml b/2.6/sitemap.xml new file mode 100644 index 0000000..0f8724e --- /dev/null +++ b/2.6/sitemap.xml @@ -0,0 +1,3 @@ + +Contributions¶
The following members contributed to the development of the CARLISLE pipeline:
- Samantha Sevilla
- Vishal Koparde
- Hsien-chao Chou
- Sohyoung Kim
- Yue Hu
- Vassiliki Saloura
VK, SS, SK, HC contributed to the generating the source code and all members contributed to the main concepts and analysis.
Overview¶
The CARLISLE github repository is stored locally, and will be used for project deployment. Multiple projects can be deployed from this one point simultaneously, without concern.
1. Getting Started¶
1.1 Introduction¶
The CARLISLE Pipelie beings with raw FASTQ files and performs trimming followed by alignment using BOWTIE2. Data is then normalized through either the use of an user-species species (IE E.Coli) spike-in control or through the determined library size. Peaks are then called using MACS2, SEACR, and GoPEAKS with various options selected by the user. Peaks are then annotated, and summarized into reports. If designated, differential analysis is performed using DESEQ2. QC reports are also generated with each project using FASTQC and MULTIQC. Annotations are added using HOMER and ROSE. GSEA Enrichment analysis predictions are added using CHIPENRICH.
The following are sub-commands used within CARLISLE:
- initialize: initalize the pipeline
- dryrun: predict the binding of peptides to any MHC molecule
- cluster: execute the pipeline on the Biowulf HPC
- local: execute a local, interactive, session
- git: execute GitHub actions
- unlock: unlock directory
- DAG: create DAG report
- report: create SNAKEMAKE report
- runtest: copies test manifests and files to WORKDIR
1.2 Setup Dependencies¶
CARLISLE has several dependencies listed below. These dependencies can be installed by a sysadmin. All dependencies will be automatically loaded if running from Biowulf.
- bedtools: "bedtools/2.30.0"
- bedops: "bedops/2.4.40"
- bowtie2: "bowtie/2-2.4.2"
- cutadapt: "cutadapt/1.18"
- fastqc: "fastqc/0.11.9"
- fastq_screen: "fastq_screen/0.15.2"
- fastq_val: "/data/CCBR_Pipeliner/iCLIP/bin/fastQValidator"
- fastxtoolkit: "fastxtoolkit/0.0.14"
- gopeaks: "github clone https://github.com/maxsonBraunLab/gopeaks"
- macs2: "macs/2.2.7.1"
- multiqc: "multiqc/1.9"
- perl: "perl/5.34.0"
- picard: "picard/2.26.9"
- python37: "python/3.7"
- R: "R/4.2.2"
- rose: "ROSE/1.3.1"
- samtools: "samtools/1.15"
- seacr: "seacr/1.4-beta.2"
- ucsc: "ucsc/407"
1.3 Login to the cluster¶
CARLISLE has been exclusively tested on Biowulf HPC. Login to the cluster's head node and move into the pipeline location.
# ssh into cluster's head node
+ssh -Y $USER@biowulf.nih.gov
+1.4 Load an interactive session¶
An interactive session should be started before performing any of the pipeline sub-commands, even if the pipeline is to be executed on the cluster.
# Grab an interactive node
+sinteractive --time=12:00:00 --mem=8gb --cpus-per-task=4 --pty bash
+4. Expected Outputs¶
The following directories are created under the WORKDIR/results directory:
- alignment_stats: this directory include information on the alignment of each sample
- bam: this directory includes BAM files, statistics on samples, statistics on spike-in controls for each sample
- bedgraph: this directory includes BEDGRAPH files and statistic summaries for each sample
- bigwig: this directory includes the bigwig files for each sample
- peaks: this directory contains a sub-directory that relates to the quality threshold used.
- quality threshold
- contrasts: this directory includes the contrasts for each line listed in the contrast manifest
- peak_caller: this directory includes all peak calls from each peak_caller (SEACR, MACS2, GOPEAKS) for each sample
- annotation
- go_enrichment: this directory includes gene set enrichment pathway predictions when
run_go_enrichmentis set totruein the config file. - homer: this directory includes the annotation output from HOMER
- rose: this directory includes the annotation output from ROSE when
run_roseis set totruein the config file.
- go_enrichment: this directory includes gene set enrichment pathway predictions when
- qc: this directory includes MULTIQC reports and spike-in control reports (when applicable)
├── alignment_stats
+├── bam
+├── bedgraph
+├── bigwig
+├── fragments
+├── peaks
+│ ├── 0.05
+│ │ ├── contrasts
+│ │ │ ├── contrast_id1.dedup_status
+│ │ │ └── contrast_id2.dedup_status
+│ │ ├── gopeaks
+│ │ │ ├── annotation
+│ │ │ │ ├── go_enrichment
+│ │ │ │ │ ├── contrast_id1.dedup_status.go_enrichment_tables
+│ │ │ │ │ └── contrast_id2.dedup_status.go_enrichment_html_report
+│ │ │ │ ├── homer
+│ │ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.gopeaks_broad.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.gopeaks_narrow.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.gopeaks_broad.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.gopeaks_narrow.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ └── rose
+│ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.gopeaks_broad.12500
+│ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.gopeaks_narrow.12500
+│ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.dedup.gopeaks_broad.12500
+│ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.dedup.gopeaks_narrow.12500
+│ │ │ └── peak_output
+│ │ ├── macs2
+│ │ │ ├── annotation
+│ │ │ │ ├── go_enrichment
+│ │ │ │ │ ├── contrast_id1.dedup_status.go_enrichment_tables
+│ │ │ │ │ └── contrast_id2.dedup_status.go_enrichment_html_report
+│ │ │ │ ├── homer
+│ │ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.macs2_narrow.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.macs2_broad.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.macs2_narrow.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.macs2_broad.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ └── rose
+│ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.macs2_broad.12500
+│ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.macs2_narrow.12500
+│ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.macs2_broad.12500
+│ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.macs2_narrow.12500
+│ │ │ └── peak_output
+│ │ └── seacr
+│ │ │ ├── annotation
+│ │ │ │ ├── go_enrichment
+│ │ │ │ │ ├── contrast_id1.dedup_status.go_enrichment_tables
+│ │ │ │ │ └── contrast_id2.dedup_status.go_enrichment_html_report
+│ │ │ │ ├── homer
+│ │ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.seacr_non_relaxed.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.seacr_non_stringent.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.seacr_norm_relaxed.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.seacr_norm_stringent.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.seacr_non_relaxed.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.seacr_non_stringent.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.seacr_norm_relaxed.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.seacr_norm_stringent.motifs
+│ │ │ │ │ │ ├── homerResults
+│ │ │ │ │ │ └── knownResults
+│ │ │ │ └── rose
+│ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.seacr_non_relaxed.12500
+│ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.seacr_non_stringent.12500
+│ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.seacr_norm_relaxed.12500
+│ │ │ │ ├── replicate_id1_vs_control_id.dedup_status.seacr_norm_stringent.12500
+│ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.seacr_non_relaxed.12500
+│ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.seacr_non_stringent.12500
+│ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.seacr_norm_relaxed.12500
+│ │ │ │ ├── replicate_id2_vs_control_id.dedup_status.seacr_norm_stringent.12500
+│ │ └── peak_output
+└── qc
+ ├── fastqc_raw
+ └── fqscreen_raw
+2. Preparing Files¶
The pipeline is controlled through editing configuration and manifest files. Defaults are found in the /WORKDIR/config and /WORKDIR/manifest directories, after initialization.
2.1 Configs¶
The configuration files control parameters and software of the pipeline. These files are listed below:
- config/config.yaml
- resources/cluster.yaml
- resources/tools.yaml
2.1.1 Cluster Config¶
The cluster configuration file dictates the resouces to be used during submission to Biowulf HPC. There are two differnt ways to control these parameters - first, to control the default settings, and second, to create or edit individual rules. These parameters should be edited with caution, after significant testing.
2.1.2 Tools Config¶
The tools configuration file dictates the version of each software or program that is being used in the pipeline.
2.1.3 Config YAML¶
There are several groups of parameters that are editable for the user to control the various aspects of the pipeline. These are :
- Folders and Paths
- These parameters will include the input and ouput files of the pipeline, as well as list all manifest names.
- User parameters
- These parameters will control the pipeline features. These include thresholds and whether to perform processes.
- References
- These parameters will control the location of index files, spike-in references, adaptors and species calling information.
2.1.3.1 User Parameters¶
2.1.3.1.1 (Spike in Controls)¶
The pipeline allows for the use of a species specific spike-in control, or the use of normalization via library size. The parameter spikein_genome should be set to the species term used in spikein_reference.
For example for ecoli spike-in:
run_contrasts: true
+norm_method: "spikein"
+spikein_genome: "ecoli"
+spikein_reference:
+ ecoli:
+ fa: "PIPELINE_HOME/resources/spikein/Ecoli_GCF_000005845.2_ASM584v2_genomic.fna"
+For example for drosophila spike-in:
run_contrasts: true
+norm_method: "spikein"
+spikein_genome: "drosophila"
+spikein_reference:
+ drosophila:
+ fa: "/fdb/igenomes/Drosophila_melanogaster/UCSC/dm6/Sequence/WholeGenomeFasta/genome.fa"
+If it's determined that the amount of spike-in is not sufficient for the run, a library normaliaztion can be performed.
- Complete a CARLISLE run with spike-in set to "Y". This will allow for the complete assessment of the spike-in.
- Run inital QC analysis on the output data
- Add the alignment_stats dir to the configuration file.
- Re-run the CARLISLE pipeline
2.1.3.1.2 Duplication Status¶
Users can select duplicated peaks (dedup) or non-deduplicated peaks (no_dedup) through the user parameter.
dupstatus: "dedup, no_dedup"
+2.1.3.1.3 Peak Caller¶
Three peak callers are available for deployment within the pipeline, with different settings deployed for each caller.
- MACS2 is available with two peak calling options: narrowPeak or broadPeak. NOTE: DESeq step generally fails for broadPeak; generally has too many calls.
peaktype: "macs2_narrow, macs2_broad,"
+- SEACR is available with four peak calling options: stringent or relaxed parameters, to be paired with "norm" for samples without a spike-in control and "non" for samples with a spikein control
peaktype: "seacr_stringent, seacr_relaxed"
+- GOPEAKS is available with two peak calling options: narrowpeaks or broadpeaks
peaktype: "gopeaks_narrow, gopeaks_broad"
+A complete list of the available peak calling parameters and the recommended list of parameters is provided below:
| Peak Caller | Narrow | Broad | Normalized, Stringent | Normalized, Relaxed | Non-Normalized, Stringent | Non-Normalized, Relaxed |
|---|---|---|---|---|---|---|
| Macs2 | AVAIL | AVAIL | NA | NA | NA | NA |
| SEACR | NA | NA | AVAIL w/o SPIKEIN | AVAIL w/o SPIKEIN | AVAIL w/ SPIKEIN | AVAIL w/ SPIKEIN |
| GoPeaks | AVAIL | AVAIL | NA | NA | NA | NA |
# Recommended list
+### peaktype: "macs2_narrow, macs2_broad, gopeaks_narrow, gopeaks_broad"
+
+# Available list
+### peaktype: "macs2_narrow, macs2_broad, seacr_norm_stringent, seacr_norm_relaxed, seacr_non_stringent, seacr_non_relaxed, gopeaks_narrow, gopeaks_broad"
+2.1.3.1.3.1 Macs2 additional option¶
MACS2 can be run with or without the control. adding a control will increase peak specificity Selecting "Y" for the macs2_control will run the paired control sample provided in the sample manifest
2.1.3.1.4 Quality Tresholds¶
Thresholds for quality can be controled through the quality_tresholds parameter. This must be a list of comma separated values. minimum of numeric value required.
- default MACS2 qvalue is 0.05 https://manpages.ubuntu.com/manpages/xenial/man1/macs2_callpeak.1.html
- default GOPEAKS pvalue is 0.05 https://github.com/maxsonBraunLab/gopeaks/blob/main/README.md
- default SEACR FDR threshold 1 https://github.com/FredHutch/SEACR/blob/master/README.md
#default values
+quality_thresholds: "0.1, 0.05, 0.01"
+2.1.3.2 References¶
Additional reference files may be added to the pipeline, if other species were to be used.
The absolute file paths which must be included are:
- fa: "/path/to/species.fa"
- blacklist: "/path/to/blacklistbed/species.bed"
The following information must be included:
- regions: "list of regions to be included; IE chr1 chr2 chr3"
- macs2_g: "macs2 genome shorthand; IE mm IE hs"
2.2 Preparing Manifests¶
There are two manifests, one which required for all pipeliens and one that is only required if running a differential analysis. These files describe information on the samples and desired contrasts. The paths of these files are defined in the snakemake_config.yaml file. These files are:
- samplemanifest
- contrasts
2.2.1 Samples Manifest (REQUIRED)¶
This manifest will include information to sample level information. It includes the following column headers:
- sampleName: the sample name WITHOUT replicate number (IE "SAMPLE")
- replicateNumber: the sample replicate number (IE "1")
- isControl: whether the sample should be identified as a control (IE "Y")
- controlName: the name of the control to use for this sample (IE "CONTROL")
- controlReplicateNumber: the replicate number of the control to use for this sample (IE "1")
- path_to_R1: the full path to R1 fastq file (IE "/path/to/sample1.R1.fastq")
- path_to_R2: the full path to R1 fastq file (IE "/path/to/sample2.R2.fastq")
An example sampleManifest file is shown below:
| sampleName | replicateNumber | isControl | controlName | controlReplicateNumber | path_to_R1 | path_to_R2 |
|---|---|---|---|---|---|---|
| 53_H3K4me3 | 1 | N | HN6_IgG_rabbit_negative_control | 1 | PIPELINE_HOME/.test/53_H3K4me3_1.R1.fastq.gz | PIPELINE_HOME/.test/53_H3K4me3_1.R2.fastq.gz |
| 53_H3K4me3 | 2 | N | HN6_IgG_rabbit_negative_control | 1 | PIPELINE_HOME/.test/53_H3K4me3_2.R1.fastq.gz | PIPELINE_HOME/.test/53_H3K4me3_2.R2.fastq.gz |
| HN6_H3K4me3 | 1 | N | HN6_IgG_rabbit_negative_control | 1 | PIPELINE_HOME/.test/HN6_H3K4me3_1.R1.fastq.gz | PIPELINE_HOME/.test/HN6_H3K4me3_1.R2.fastq.gz |
| HN6_H3K4me3 | 2 | N | HN6_IgG_rabbit_negative_control | 1 | PIPELINE_HOME/.test/HN6_H3K4me3_2.R1.fastq.gz | PIPELINE_HOME/.test/HN6_H3K4me3_2.R2.fastq.gz |
| HN6_IgG_rabbit_negative_control | 1 | Y | - | - | PIPELINE_HOME/.test/HN6_IgG_rabbit_negative_control_1.R1.fastq.gz | PIPELINE_HOME/.test/HN6_IgG_rabbit_negative_control_1.R2.fastq.gz |
2.2.2 Contrast Manifest (OPTIONAL)¶
This manifest will include sample information to performed differential comparisons.
An example contrast file:
| condition1 | condition2 |
|---|---|
| MOC1_siSmyd3_2m_25_HCHO | MOC1_siNC_2m_25_HCHO |
Note: you must have more than one sample per condition in order to perform differential analysis with DESeq2
3. Running the Pipeline¶
3.1 Pipeline Overview¶
The Snakemake workflow has a multiple options
Required arguments¶
Usage: bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle -m/--runmode=<RUNMODE> -w/--workdir=<WORKDIR>
+
+1. RUNMODE: [Type: String] Valid options:
+ *) init : initialize workdir
+ *) run : run with slurm
+ *) reset : DELETE workdir dir and re-init it
+ *) dryrun : dry run snakemake to generate DAG
+ *) unlock : unlock workdir if locked by snakemake
+ *) runlocal : run without submitting to sbatch
+ *) runtest: run on cluster with included test dataset
+2. WORKDIR: [Type: String]: Absolute or relative path to the output folder with write permissions.
+Optional arguments¶
--help|-h : print this help. --version|-v : print the version of carlisle. --force|-f : use the force flag for snakemake to force all rules to run. --singcache|-c : singularity cache directory. Default is /data/${USER}/.singularity if available, or falls back to ${WORKDIR}/.singularity. Use this flag to specify a different singularity cache directory.
3.2 Commands explained¶
The following explains each of the command options:
- Preparation Commands
- init (REQUIRED): This must be performed before any Snakemake run (dry, local, cluster) can be performed. This will copy the necessary config, manifest and Snakefiles needed to run the pipeline to the provided output directory.
- the -f/--force flag can be used in order to re-initialize a workdir that has already been created
- dryrun (OPTIONAL): This is an optional step, to be performed before any Snakemake run (local, cluster). This will check for errors within the pipeline, and ensure that you have read/write access to the files needed to run the full pipeline.
- Processing Commands
- local: This will run the pipeline on a local node. NOTE: This should only be performed on an interactive node.
- run: This will submit a master job to the cluster, and subsequent sub-jobs as needed to complete the workflow. An email will be sent when the pipeline begins, if there are any errors, and when it completes.
- Other Commands (All optional)
- unlock: This will unlock the pipeline if an error caused it to stop in the middle of a run.
- runtest: This will run a test of the pipeline with test data
To run any of these commands, follow the the syntax:
bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=COMMAND --workdir=/path/to/output/dir
+3.3 Typical Workflow¶
A typical command workflow, running on the cluser, is as follows:
bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=init --workdir=/path/to/output/dir
+
+bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=dryrun --workdir=/path/to/output/dir
+
+bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=run --workdir=/path/to/output/dir
+5. Pipeline Tutorial¶
Welcome to the CARLISLE Pipeline Tutorial!
5.1 Getting Started¶
Review the information on the Getting Started for a complete overview the pipeline. The tutorial below will use test data available on NIH Biowulf HPC only. All example code will assume you are running v1.0 of the pipeline, using test data available on GitHub.
A. Change working directory to the CARLISLE repository
B. Initialize Pipeline
bash ./path/to/dir/carlisle --runmode=init --workdir=/path/to/output/dir
+5.2 Submit the test data¶
Test data is included in the .test directory as well as the config directory.
A Run the test command to prepare the data, perform a dry-run and submit to the cluster
bash ./path/to/dir/carlisle --runmode=runtest --workdir=/path/to/output/dir
+- An expected output for the
runtestis as follows:
Job stats:
+job count min threads max threads
+----------------------------- ------- ------------- -------------
+DESeq 24 1 1
+align 9 56 56
+alignstats 9 2 2
+all 1 1 1
+bam2bg 9 4 4
+create_contrast_data_files 24 1 1
+create_contrast_peakcaller_files 12 1 1
+create_reference 1 32 32
+create_replicate_sample_table 1 1 1
+diffbb 24 1 1
+filter 18 2 2
+findMotif 96 6 6
+gather_alignstats 1 1 1
+go_enrichment 12 1 1
+gopeaks_broad 16 2 2
+gopeaks_narrow 16 2 2
+macs2_broad 16 2 2
+macs2_narrow 16 2 2
+make_counts_matrix 24 1 1
+multiqc 2 1 1
+qc_fastqc 9 1 1
+rose 96 2 2
+seacr_relaxed 16 2 2
+seacr_stringent 16 2 2
+spikein_assessment 1 1 1
+trim 9 56 56
+total 478 1 56
+5.3 Review outputs¶
Review the expected outputs on the Output page. If there are errors, review and performing stesp described on the Troubleshooting page as needed.
Troubleshooting¶
Recommended steps to troubleshoot the pipeline.
1.1 Email¶
Check your email for an email regarding pipeline failure. You will receive an email from slurm@biowulf.nih.gov with the subject: Slurm Job_id=[#] Name=CARLISLE Failed, Run time [time], FAILED, ExitCode 1
1.2 Review the log files¶
Review the logs in two ways:
- Review the master slurm file: This file will be found in the
/path/to/results/dir/and titledslurm-[jobid].out. Reviewing this file will tell you what rule errored, and for any local SLURM jobs, provide error details - Review the individual rule log files: After reviewing the master slurm-file, review the specific rules that failed within the
/path/to/results/dir/logs/. Each rule will include a.errand.outfile, with the following formatting:{rulename}.{masterjobID}.{individualruleID}.{wildcards from the rule}.{out or err}
1.3 Restart the run¶
After addressing the issue, unlock the output directory, perform another dry-run and check the status of the pipeline, then resubmit to the cluster.
#unlock dir
+bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=unlock --workdir=/path/to/output/dir
+
+#perform dry-run
+bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=dryrun --workdir=/path/to/output/dir
+
+#submit to cluster
+bash ./data/CCBR_Pipeliner/Pipelines/CARLISLE/carlisle --runmode=run --workdir=/path/to/output/dir
+1.4 Contact information¶
If after troubleshooting, the error cannot be resolved, or if a bug is found, please create an issue and send and email to Samantha Chill.


